Hung-Chun Hsu
CFDA & CLIP at TREC iKAT 2025: Enhancing Personalized Conversational Search via Query Reformulation and Rank Fusion
Sep 19, 2025Abstract:The 2025 TREC Interactive Knowledge Assistance Track (iKAT) featured both interactive and offline submission tasks. The former requires systems to operate under real-time constraints, making robustness and efficiency as important as accuracy, while the latter enables controlled evaluation of passage ranking and response generation with pre-defined datasets. To address this, we explored query rewriting and retrieval fusion as core strategies. We built our pipelines around Best-of-$N$ selection and Reciprocal Rank Fusion (RRF) strategies to handle different submission tasks. Results show that reranking and fusion improve robustness while revealing trade-offs between effectiveness and efficiency across both tasks.
Test-Time Scaling Strategies for Generative Retrieval in Multimodal Conversational Recommendations
Aug 25, 2025



Abstract:The rapid evolution of e-commerce has exposed the limitations of traditional product retrieval systems in managing complex, multi-turn user interactions. Recent advances in multimodal generative retrieval -- particularly those leveraging multimodal large language models (MLLMs) as retrievers -- have shown promise. However, most existing methods are tailored to single-turn scenarios and struggle to model the evolving intent and iterative nature of multi-turn dialogues when applied naively. Concurrently, test-time scaling has emerged as a powerful paradigm for improving large language model (LLM) performance through iterative inference-time refinement. Yet, its effectiveness typically relies on two conditions: (1) a well-defined problem space (e.g., mathematical reasoning), and (2) the model's ability to self-correct -- conditions that are rarely met in conversational product search. In this setting, user queries are often ambiguous and evolving, and MLLMs alone have difficulty grounding responses in a fixed product corpus. Motivated by these challenges, we propose a novel framework that introduces test-time scaling into conversational multimodal product retrieval. Our approach builds on a generative retriever, further augmented with a test-time reranking (TTR) mechanism that improves retrieval accuracy and better aligns results with evolving user intent throughout the dialogue. Experiments across multiple benchmarks show consistent improvements, with average gains of 14.5 points in MRR and 10.6 points in nDCG@1.
NINEPINS: Nuclei Instance Segmentation with Point Annotations
Jun 24, 2020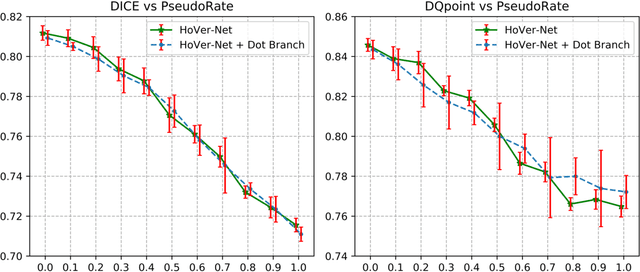
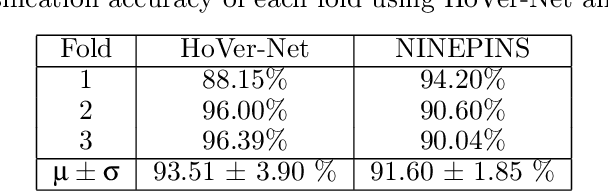
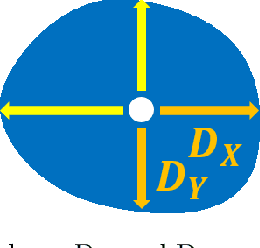
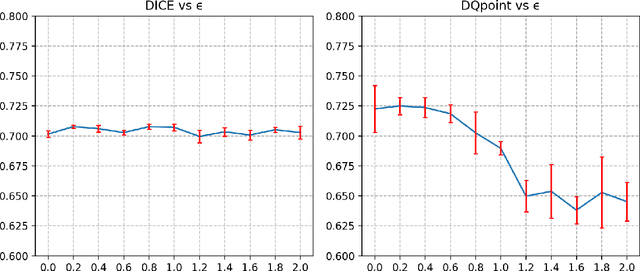
Abstract:Deep learning-based methods are gaining traction in digital pathology, with an increasing number of publications and challenges that aim at easing the work of systematically and exhaustively analyzing tissue slides. These methods often achieve very high accuracies, at the cost of requiring large annotated datasets to train. This requirement is especially difficult to fulfill in the medical field, where expert knowledge is essential. In this paper we focus on nuclei segmentation, which generally requires experienced pathologists to annotate the nuclear areas in gigapixel histological images. We propose an algorithm for instance segmentation that uses pseudo-label segmentations generated automatically from point annotations, as a method to reduce the burden for pathologists. With the generated segmentation masks, the proposed method trains a modified version of HoVer-Net model to achieve instance segmentation. Experimental results show that the proposed method is robust to inaccuracies in point annotations and comparison with Hover-Net trained with fully annotated instance masks shows that a degradation in segmentation performance does not always imply a degradation in higher order tasks such as tissue classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge