Hritam Basak
UA-Pose: Uncertainty-Aware 6D Object Pose Estimation and Online Object Completion with Partial References
Jun 09, 2025



Abstract:6D object pose estimation has shown strong generalizability to novel objects. However, existing methods often require either a complete, well-reconstructed 3D model or numerous reference images that fully cover the object. Estimating 6D poses from partial references, which capture only fragments of an object's appearance and geometry, remains challenging. To address this, we propose UA-Pose, an uncertainty-aware approach for 6D object pose estimation and online object completion specifically designed for partial references. We assume access to either (1) a limited set of RGBD images with known poses or (2) a single 2D image. For the first case, we initialize a partial object 3D model based on the provided images and poses, while for the second, we use image-to-3D techniques to generate an initial object 3D model. Our method integrates uncertainty into the incomplete 3D model, distinguishing between seen and unseen regions. This uncertainty enables confidence assessment in pose estimation and guides an uncertainty-aware sampling strategy for online object completion, enhancing robustness in pose estimation accuracy and improving object completeness. We evaluate our method on the YCB-Video, YCBInEOAT, and HO3D datasets, including RGBD sequences of YCB objects manipulated by robots and human hands. Experimental results demonstrate significant performance improvements over existing methods, particularly when object observations are incomplete or partially captured. Project page: https://minfenli.github.io/UA-Pose/
SemiDAViL: Semi-supervised Domain Adaptation with Vision-Language Guidance for Semantic Segmentation
Apr 08, 2025



Abstract:Domain Adaptation (DA) and Semi-supervised Learning (SSL) converge in Semi-supervised Domain Adaptation (SSDA), where the objective is to transfer knowledge from a source domain to a target domain using a combination of limited labeled target samples and abundant unlabeled target data. Although intuitive, a simple amalgamation of DA and SSL is suboptimal in semantic segmentation due to two major reasons: (1) previous methods, while able to learn good segmentation boundaries, are prone to confuse classes with similar visual appearance due to limited supervision; and (2) skewed and imbalanced training data distribution preferring source representation learning whereas impeding from exploring limited information about tailed classes. Language guidance can serve as a pivotal semantic bridge, facilitating robust class discrimination and mitigating visual ambiguities by leveraging the rich semantic relationships encoded in pre-trained language models to enhance feature representations across domains. Therefore, we propose the first language-guided SSDA setting for semantic segmentation in this work. Specifically, we harness the semantic generalization capabilities inherent in vision-language models (VLMs) to establish a synergistic framework within the SSDA paradigm. To address the inherent class-imbalance challenges in long-tailed distributions, we introduce class-balanced segmentation loss formulations that effectively regularize the learning process. Through extensive experimentation across diverse domain adaptation scenarios, our approach demonstrates substantial performance improvements over contemporary state-of-the-art (SoTA) methodologies. Code is available: \href{https://github.com/hritam-98/SemiDAViL}{GitHub}.
Enhancing Single Image to 3D Generation using Gaussian Splatting and Hybrid Diffusion Priors
Oct 12, 2024



Abstract:3D object generation from a single image involves estimating the full 3D geometry and texture of unseen views from an unposed RGB image captured in the wild. Accurately reconstructing an object's complete 3D structure and texture has numerous applications in real-world scenarios, including robotic manipulation, grasping, 3D scene understanding, and AR/VR. Recent advancements in 3D object generation have introduced techniques that reconstruct an object's 3D shape and texture by optimizing the efficient representation of Gaussian Splatting, guided by pre-trained 2D or 3D diffusion models. However, a notable disparity exists between the training datasets of these models, leading to distinct differences in their outputs. While 2D models generate highly detailed visuals, they lack cross-view consistency in geometry and texture. In contrast, 3D models ensure consistency across different views but often result in overly smooth textures. We propose bridging the gap between 2D and 3D diffusion models to address this limitation by integrating a two-stage frequency-based distillation loss with Gaussian Splatting. Specifically, we leverage geometric priors in the low-frequency spectrum from a 3D diffusion model to maintain consistent geometry and use a 2D diffusion model to refine the fidelity and texture in the high-frequency spectrum of the generated 3D structure, resulting in more detailed and fine-grained outcomes. Our approach enhances geometric consistency and visual quality, outperforming the current SOTA. Additionally, we demonstrate the easy adaptability of our method for efficient object pose estimation and tracking.
Semi-supervised Domain Adaptive Medical Image Segmentation through Consistency Regularized Disentangled Contrastive Learning
Jul 06, 2023Abstract:Although unsupervised domain adaptation (UDA) is a promising direction to alleviate domain shift, they fall short of their supervised counterparts. In this work, we investigate relatively less explored semi-supervised domain adaptation (SSDA) for medical image segmentation, where access to a few labeled target samples can improve the adaptation performance substantially. Specifically, we propose a two-stage training process. First, an encoder is pre-trained in a self-learning paradigm using a novel domain-content disentangled contrastive learning (CL) along with a pixel-level feature consistency constraint. The proposed CL enforces the encoder to learn discriminative content-specific but domain-invariant semantics on a global scale from the source and target images, whereas consistency regularization enforces the mining of local pixel-level information by maintaining spatial sensitivity. This pre-trained encoder, along with a decoder, is further fine-tuned for the downstream task, (i.e. pixel-level segmentation) using a semi-supervised setting. Furthermore, we experimentally validate that our proposed method can easily be extended for UDA settings, adding to the superiority of the proposed strategy. Upon evaluation on two domain adaptive image segmentation tasks, our proposed method outperforms the SoTA methods, both in SSDA and UDA settings. Code is available at https://github.com/hritam-98/GFDA-disentangled
UT-Net: Combining U-Net and Transformer for Joint Optic Disc and Cup Segmentation and Glaucoma Detection
Mar 08, 2023



Abstract:Glaucoma is a chronic visual disease that may cause permanent irreversible blindness. Measurement of the cup-to-disc ratio (CDR) plays a pivotal role in the detection of glaucoma in its early stage, preventing visual disparities. Therefore, accurate and automatic segmentation of optic disc (OD) and optic cup (OC) from retinal fundus images is a fundamental requirement. Existing CNN-based segmentation frameworks resort to building deep encoders with aggressive downsampling layers, which suffer from a general limitation on modeling explicit long-range dependency. To this end, in this paper, we propose a new segmentation pipeline, called UT-Net, availing the advantages of U-Net and transformer both in its encoding layer, followed by an attention-gated bilinear fusion scheme. In addition to this, we incorporate Multi-Head Contextual attention to enhance the regular self-attention used in traditional vision transformers. Thus low-level features along with global dependencies are captured in a shallow manner. Besides, we extract context information at multiple encoding layers for better exploration of receptive fields, and to aid the model to learn deep hierarchical representations. Finally, an enhanced mixing loss is proposed to tightly supervise the overall learning process. The proposed model has been implemented for joint OD and OC segmentation on three publicly available datasets: DRISHTI-GS, RIM-ONE R3, and REFUGE. Additionally, to validate our proposal, we have performed exhaustive experimentation on Glaucoma detection from all three datasets by measuring the Cup to Disc Ratio (CDR) value. Experimental results demonstrate the superiority of UT-Net as compared to the state-of-the-art methods.
IDEAL: Improved DEnse locAL Contrastive Learning for Semi-Supervised Medical Image Segmentation
Oct 26, 2022



Abstract:Due to the scarcity of labeled data, Contrastive Self-Supervised Learning (SSL) frameworks have lately shown great potential in several medical image analysis tasks. However, the existing contrastive mechanisms are sub-optimal for dense pixel-level segmentation tasks due to their inability to mine local features. To this end, we extend the concept of metric learning to the segmentation task, using a dense (dis)similarity learning for pre-training a deep encoder network, and employing a semi-supervised paradigm to fine-tune for the downstream task. Specifically, we propose a simple convolutional projection head for obtaining dense pixel-level features, and a new contrastive loss to utilize these dense projections thereby improving the local representations. A bidirectional consistency regularization mechanism involving two-stream model training is devised for the downstream task. Upon comparison, our IDEAL method outperforms the SoTA methods by fair margins on cardiac MRI segmentation.
MFSNet: A Multi Focus Segmentation Network for Skin Lesion Segmentation
Mar 29, 2022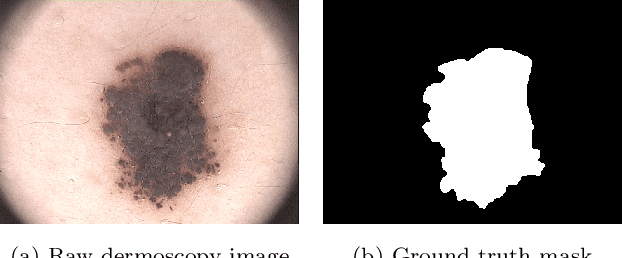
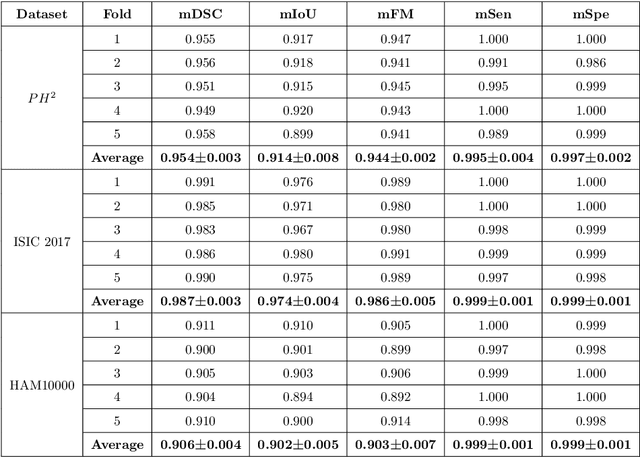
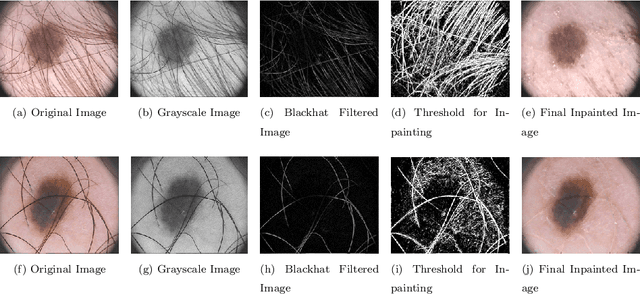
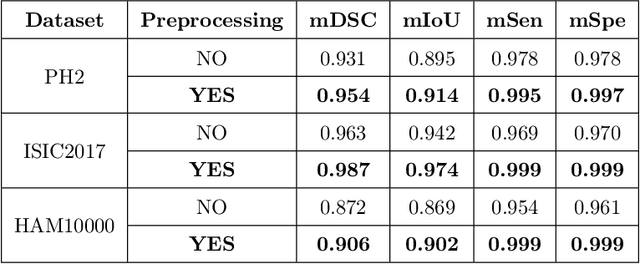
Abstract:Segmentation is essential for medical image analysis to identify and localize diseases, monitor morphological changes, and extract discriminative features for further diagnosis. Skin cancer is one of the most common types of cancer globally, and its early diagnosis is pivotal for the complete elimination of malignant tumors from the body. This research develops an Artificial Intelligence (AI) framework for supervised skin lesion segmentation employing the deep learning approach. The proposed framework, called MFSNet (Multi-Focus Segmentation Network), uses differently scaled feature maps for computing the final segmentation mask using raw input RGB images of skin lesions. In doing so, initially, the images are preprocessed to remove unwanted artifacts and noises. The MFSNet employs the Res2Net backbone, a recently proposed convolutional neural network (CNN), for obtaining deep features used in a Parallel Partial Decoder (PPD) module to get a global map of the segmentation mask. In different stages of the network, convolution features and multi-scale maps are used in two boundary attention (BA) modules and two reverse attention (RA) modules to generate the final segmentation output. MFSNet, when evaluated on three publicly available datasets: $PH^2$, ISIC 2017, and HAM10000, outperforms state-of-the-art methods, justifying the reliability of the framework. The relevant codes for the proposed approach are accessible at https://github.com/Rohit-Kundu/MFSNet
An Embarrassingly Simple Consistency Regularization Method for Semi-Supervised Medical Image Segmentation
Feb 03, 2022
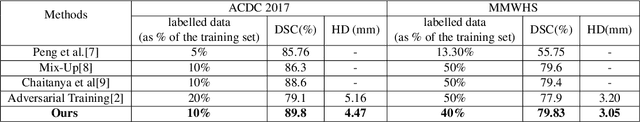
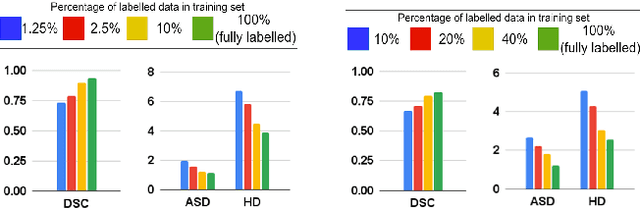
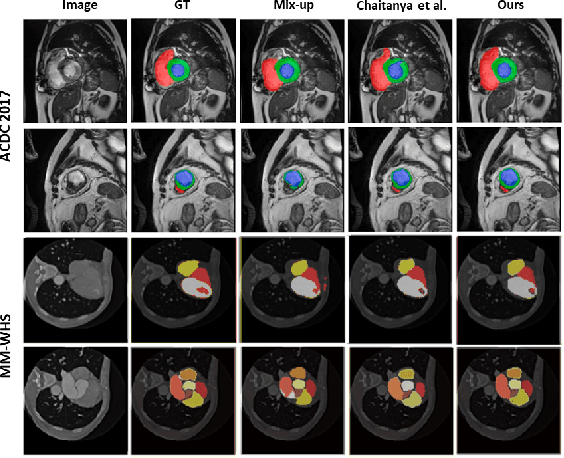
Abstract:The scarcity of pixel-level annotation is a prevalent problem in medical image segmentation tasks. In this paper, we introduce a novel regularization strategy involving interpolation-based mixing for semi-supervised medical image segmentation. The proposed method is a new consistency regularization strategy that encourages segmentation of interpolation of two unlabelled data to be consistent with the interpolation of segmentation maps of those data. This method represents a specific type of data-adaptive regularization paradigm which aids to minimize the overfitting of labelled data under high confidence values. The proposed method is advantageous over adversarial and generative models as it requires no additional computation. Upon evaluation on two publicly available MRI datasets: ACDC and MMWHS, experimental results demonstrate the superiority of the proposed method in comparison to existing semi-supervised models. Code is available at: https://github.com/hritam-98/ICT-MedSeg
SVM and ANN based Classification of EMG signals by using PCA and LDA
Oct 22, 2021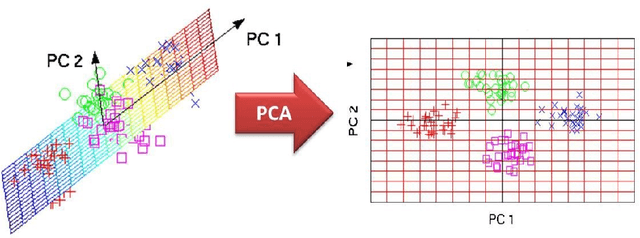
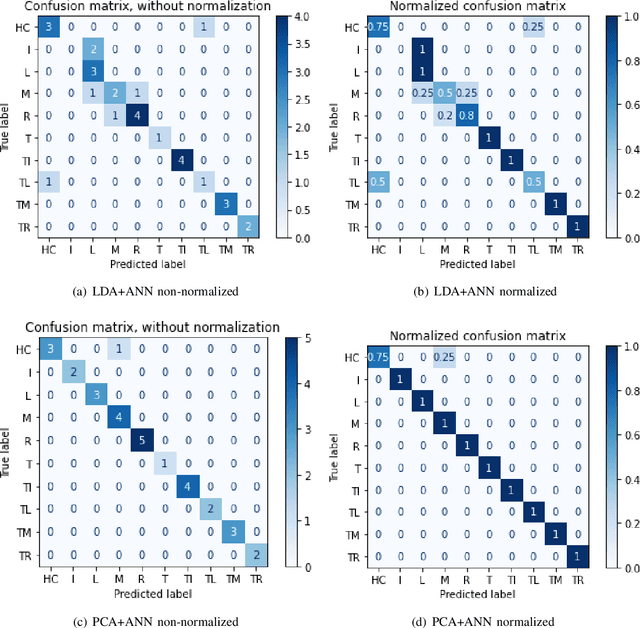
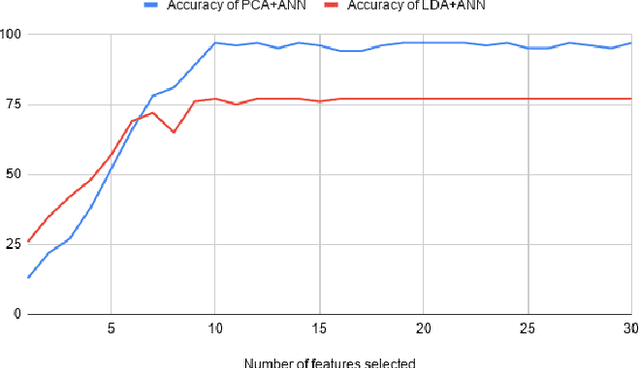
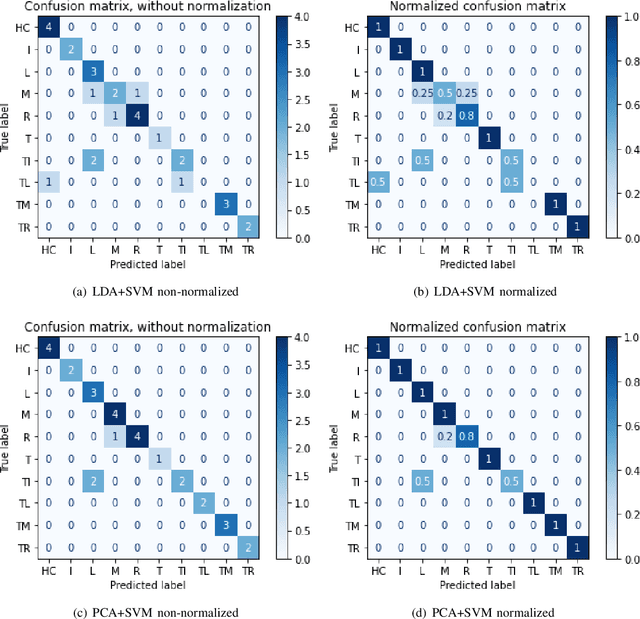
Abstract:In recent decades, biomedical signals have been used for communication in Human-Computer Interfaces (HCI) for medical applications; an instance of these signals are the myoelectric signals (MES), which are generated in the muscles of the human body as unidimensional patterns. Because of this, the methods and algorithms developed for pattern recognition in signals can be applied for their analyses once these signals have been sampled and turned into electromyographic (EMG) signals. Additionally, in recent years, many researchers have dedicated their efforts to studying prosthetic control utilizing EMG signal classification, that is, by logging a set of MES in a proper range of frequencies to classify the corresponding EMG signals. The feature classification can be carried out on the time domain or by using other domains such as the frequency domain (also known as the spectral domain), time scale, and time-frequency, amongst others. One of the main methods used for pattern recognition in myoelectric signals is the Support Vector Machines (SVM) technique whose primary function is to identify an n-dimensional hyperplane to separate a set of input feature points into different classes. This technique has the potential to recognize complex patterns and on several occasions, it has proven its worth when compared to other classifiers such as Artificial Neural Network (ANN), Linear Discriminant Analysis (LDA), and Principal Component Analysis(PCA). The key concepts underlying the SVM are (a) the hyperplane separator; (b) the kernel function; (c) the optimal separation hyperplane; and (d) a soft margin (hyperplane tolerance).
Ensemble of CNN classifiers using Sugeno Fuzzy Integral Technique for Cervical Cytology Image Classification
Aug 21, 2021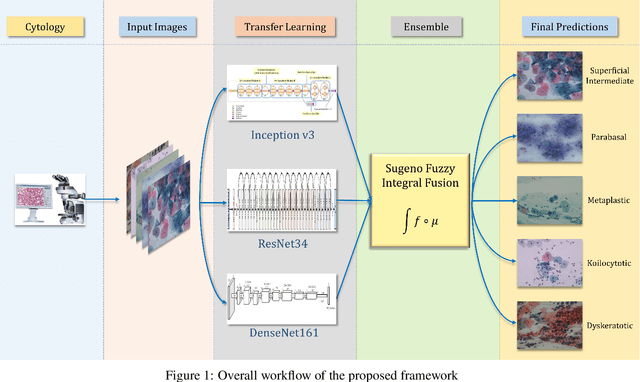
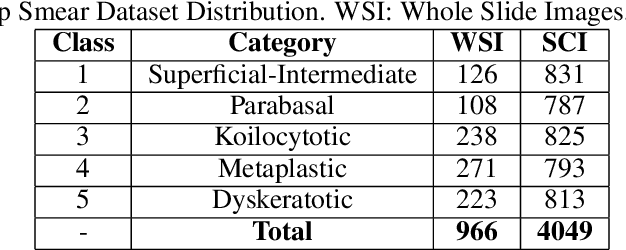
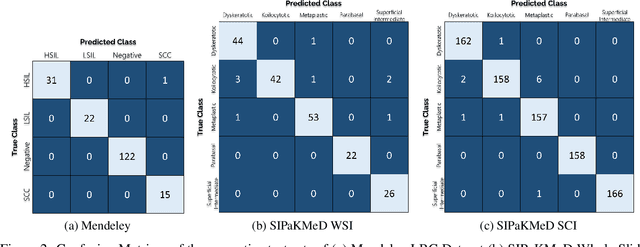
Abstract:Cervical cancer is the fourth most common category of cancer, affecting more than 500,000 women annually, owing to the slow detection procedure. Early diagnosis can help in treating and even curing cancer, but the tedious, time-consuming testing process makes it impossible to conduct population-wise screening. To aid the pathologists in efficient and reliable detection, in this paper, we propose a fully automated computer-aided diagnosis tool for classifying single-cell and slide images of cervical cancer. The main concern in developing an automatic detection tool for biomedical image classification is the low availability of publicly accessible data. Ensemble Learning is a popular approach for image classification, but simplistic approaches that leverage pre-determined weights to classifiers fail to perform satisfactorily. In this research, we use the Sugeno Fuzzy Integral to ensemble the decision scores from three popular pretrained deep learning models, namely, Inception v3, DenseNet-161 and ResNet-34. The proposed Fuzzy fusion is capable of taking into consideration the confidence scores of the classifiers for each sample, and thus adaptively changing the importance given to each classifier, capturing the complementary information supplied by each, thus leading to superior classification performance. We evaluated the proposed method on three publicly available datasets, the Mendeley Liquid Based Cytology (LBC) dataset, the SIPaKMeD Whole Slide Image (WSI) dataset, and the SIPaKMeD Single Cell Image (SCI) dataset, and the results thus yielded are promising. Analysis of the approach using GradCAM-based visual representations and statistical tests, and comparison of the method with existing and baseline models in literature justify the efficacy of the approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge