Hairong Wang
CLARiTy: A Vision Transformer for Multi-Label Classification and Weakly-Supervised Localization of Chest X-ray Pathologies
Dec 18, 2025Abstract:The interpretation of chest X-rays (CXRs) poses significant challenges, particularly in achieving accurate multi-label pathology classification and spatial localization. These tasks demand different levels of annotation granularity but are frequently constrained by the scarcity of region-level (dense) annotations. We introduce CLARiTy (Class Localizing and Attention Refining Image Transformer), a vision transformer-based model for joint multi-label classification and weakly-supervised localization of thoracic pathologies. CLARiTy employs multiple class-specific tokens to generate discriminative attention maps, and a SegmentCAM module for foreground segmentation and background suppression using explicit anatomical priors. Trained on image-level labels from the NIH ChestX-ray14 dataset, it leverages distillation from a ConvNeXtV2 teacher for efficiency. Evaluated on the official NIH split, the CLARiTy-S-16-512 (a configuration of CLARiTy), achieves competitive classification performance across 14 pathologies, and state-of-the-art weakly-supervised localization performance on 8 pathologies, outperforming prior methods by 50.7%. In particular, pronounced gains occur for small pathologies like nodules and masses. The lower-resolution variant of CLARiTy, CLARiTy-S-16-224, offers high efficiency while decisively surpassing baselines, thereby having the potential for use in low-resource settings. An ablation study confirms contributions of SegmentCAM, DINO pretraining, orthogonal class token loss, and attention pooling. CLARiTy advances beyond CNN-ViT hybrids by harnessing ViT self-attention for global context and class-specific localization, refined through convolutional background suppression for precise, noise-reduced heatmaps.
BiCoRec: Bias-Mitigated Context-Aware Sequential Recommendation Model
Dec 15, 2025Abstract:Sequential recommendation models aim to learn from users evolving preferences. However, current state-of-the-art models suffer from an inherent popularity bias. This study developed a novel framework, BiCoRec, that adaptively accommodates users changing preferences for popular and niche items. Our approach leverages a co-attention mechanism to obtain a popularity-weighted user sequence representation, facilitating more accurate predictions. We then present a new training scheme that learns from future preferences using a consistency loss function. BiCoRec aimed to improve the recommendation performance of users who preferred niche items. For these users, BiCoRec achieves a 26.00% average improvement in NDCG@10 over state-of-the-art baselines. When ranking the relevant item against the entire collection, BiCoRec achieves NDCG@10 scores of 0.0102, 0.0047, 0.0021, and 0.0005 for the Movies, Fashion, Games and Music datasets.
BrainNormalizer: Anatomy-Informed Pseudo-Healthy Brain Reconstruction from Tumor MRI via Edge-Guided ControlNet
Nov 17, 2025Abstract:Brain tumors are among the most clinically significant neurological diseases and remain a major cause of morbidity and mortality due to their aggressive growth and structural heterogeneity. As tumors expand, they induce substantial anatomical deformation that disrupts both local tissue organization and global brain architecture, complicating diagnosis, treatment planning, and surgical navigation. Yet a subject-specific reference of how the brain would appear without tumor-induced changes is fundamentally unobtainable in clinical practice. We present BrainNormalizer, an anatomy-informed diffusion framework that reconstructs pseudo-healthy MRIs directly from tumorous scans by conditioning the generative process on boundary cues extracted from the subject's own anatomy. This boundary-guided conditioning enables anatomically plausible pseudo-healthy reconstruction without requiring paired non-tumorous and tumorous scans. BrainNormalizer employs a two-stage training strategy. The pretrained diffusion model is first adapted through inpainting-based fine-tuning on tumorous and non-tumorous scans. Next, an edge-map-guided ControlNet branch is trained to inject fine-grained anatomical contours into the frozen decoder while preserving learned priors. During inference, a deliberate misalignment strategy pairs tumorous inputs with non-tumorous prompts and mirrored contralateral edge maps, leveraging hemispheric correspondence to guide reconstruction. On the BraTS2020 dataset, BrainNormalizer achieves strong quantitative performance and qualitatively produces anatomically plausible reconstructions in tumor-affected regions while retaining overall structural coherence. BrainNormalizer provides clinically reliable anatomical references for treatment planning and supports new research directions in counterfactual modeling and tumor-induced deformation analysis.
A Holistic Weakly Supervised Approach for Liver Tumor Segmentation with Clinical Knowledge-Informed Label Smoothing
Oct 13, 2024



Abstract:Liver cancer is a leading cause of mortality worldwide, and accurate CT-based tumor segmentation is essential for diagnosis and treatment. Manual delineation is time-intensive, prone to variability, and highlights the need for reliable automation. While deep learning has shown promise for automated liver segmentation, precise liver tumor segmentation remains challenging due to the heterogeneous nature of tumors, imprecise tumor margins, and limited labeled data. We present a novel holistic weakly supervised framework that integrates clinical knowledge to address these challenges with (1) A knowledge-informed label smoothing technique that leverages clinical data to generate smooth labels, which regularizes model training reducing the risk of overfitting and enhancing model performance; (2) A global and local-view segmentation framework, breaking down the task into two simpler sub-tasks, allowing optimized preprocessing and training for each; and (3) Pre- and post-processing pipelines customized to the challenges of each subtask, which enhances tumor visibility and refines tumor boundaries. We evaluated the proposed method on the HCC-TACE-Seg dataset and showed that these three key components complementarily contribute to the improved performance. Lastly, we prototyped a tool for automated liver tumor segmentation and diagnosis summary generation called MedAssistLiver. The app and code are published at https://github.com/lingchm/medassist-liver-cancer.
Knowledge-Informed Machine Learning for Cancer Diagnosis and Prognosis: A review
Jan 12, 2024



Abstract:Cancer remains one of the most challenging diseases to treat in the medical field. Machine learning has enabled in-depth analysis of rich multi-omics profiles and medical imaging for cancer diagnosis and prognosis. Despite these advancements, machine learning models face challenges stemming from limited labeled sample sizes, the intricate interplay of high-dimensionality data types, the inherent heterogeneity observed among patients and within tumors, and concerns about interpretability and consistency with existing biomedical knowledge. One approach to surmount these challenges is to integrate biomedical knowledge into data-driven models, which has proven potential to improve the accuracy, robustness, and interpretability of model results. Here, we review the state-of-the-art machine learning studies that adopted the fusion of biomedical knowledge and data, termed knowledge-informed machine learning, for cancer diagnosis and prognosis. Emphasizing the properties inherent in four primary data types including clinical, imaging, molecular, and treatment data, we highlight modeling considerations relevant to these contexts. We provide an overview of diverse forms of knowledge representation and current strategies of knowledge integration into machine learning pipelines with concrete examples. We conclude the review article by discussing future directions to advance cancer research through knowledge-informed machine learning.
Quantifying intra-tumoral genetic heterogeneity of glioblastoma toward precision medicine using MRI and a data-inclusive machine learning algorithm
Dec 30, 2023Abstract:Glioblastoma (GBM) is one of the most aggressive and lethal human cancers. Intra-tumoral genetic heterogeneity poses a significant challenge for treatment. Biopsy is invasive, which motivates the development of non-invasive, MRI-based machine learning (ML) models to quantify intra-tumoral genetic heterogeneity for each patient. This capability holds great promise for enabling better therapeutic selection to improve patient outcomes. We proposed a novel Weakly Supervised Ordinal Support Vector Machine (WSO-SVM) to predict regional genetic alteration status within each GBM tumor using MRI. WSO-SVM was applied to a unique dataset of 318 image-localized biopsies with spatially matched multiparametric MRI from 74 GBM patients. The model was trained to predict the regional genetic alteration of three GBM driver genes (EGFR, PDGFRA, and PTEN) based on features extracted from the corresponding region of five MRI contrast images. For comparison, a variety of existing ML algorithms were also applied. The classification accuracy of each gene was compared between the different algorithms. The SHapley Additive exPlanations (SHAP) method was further applied to compute contribution scores of different contrast images. Finally, the trained WSO-SVM was used to generate prediction maps within the tumoral area of each patient to help visualize the intra-tumoral genetic heterogeneity. This study demonstrated the feasibility of using MRI and WSO-SVM to enable non-invasive prediction of intra-tumoral regional genetic alteration for each GBM patient, which can inform future adaptive therapies for individualized oncology.
A Novel Hybrid Ordinal Learning Model with Health Care Application
Dec 15, 2023



Abstract:Ordinal learning (OL) is a type of machine learning models with broad utility in health care applications such as diagnosis of different grades of a disease (e.g., mild, modest, severe) and prediction of the speed of disease progression (e.g., very fast, fast, moderate, slow). This paper aims to tackle a situation when precisely labeled samples are limited in the training set due to cost or availability constraints, whereas there could be an abundance of samples with imprecise labels. We focus on imprecise labels that are intervals, i.e., one can know that a sample belongs to an interval of labels but cannot know which unique label it has. This situation is quite common in health care datasets due to limitations of the diagnostic instrument, sparse clinical visits, or/and patient dropout. Limited research has been done to develop OL models with imprecise/interval labels. We propose a new Hybrid Ordinal Learner (HOL) to integrate samples with both precise and interval labels to train a robust OL model. We also develop a tractable and efficient optimization algorithm to solve the HOL formulation. We compare HOL with several recently developed OL methods on four benchmarking datasets, which demonstrate the superior performance of HOL. Finally, we apply HOL to a real-world dataset for predicting the speed of progressing to Alzheimer's Disease (AD) for individuals with Mild Cognitive Impairment (MCI) based on a combination of multi-modality neuroimaging and demographic/clinical datasets. HOL achieves high accuracy in the prediction and outperforms existing methods. The capability of accurately predicting the speed of progression to AD for each individual with MCI has the potential for helping facilitate more individually-optimized interventional strategies.
Translated Skip Connections -- Expanding the Receptive Fields of Fully Convolutional Neural Networks
Nov 03, 2022Abstract:The effective receptive field of a fully convolutional neural network is an important consideration when designing an architecture, as it defines the portion of the input visible to each convolutional kernel. We propose a neural network module, extending traditional skip connections, called the translated skip connection. Translated skip connections geometrically increase the receptive field of an architecture with negligible impact on both the size of the parameter space and computational complexity. By embedding translated skip connections into a benchmark architecture, we demonstrate that our module matches or outperforms four other approaches to expanding the effective receptive fields of fully convolutional neural networks. We confirm this result across five contemporary image segmentation datasets from disparate domains, including the detection of COVID-19 infection, segmentation of aerial imagery, common object segmentation, and segmentation for self-driving cars.
* 5 pages, 2 figures, 1 table, published at the 2022 IEEE International Conference on Image Processing
Multi-Modal Recommendation System with Auxiliary Information
Oct 13, 2022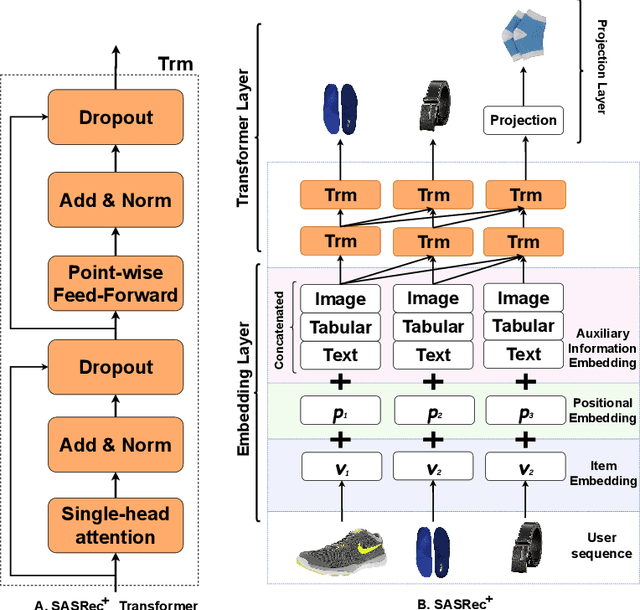
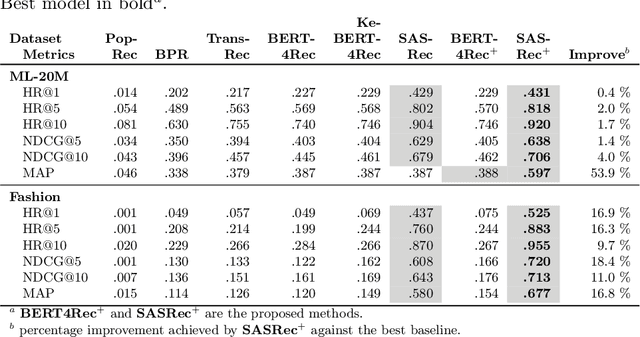
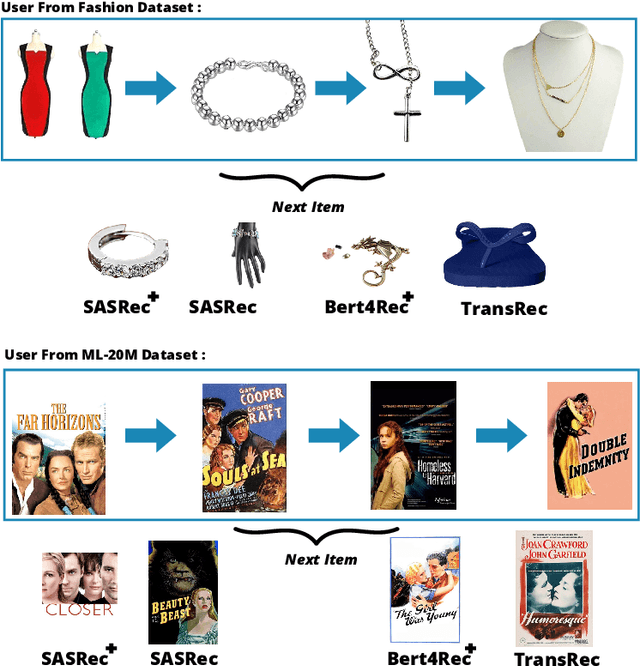
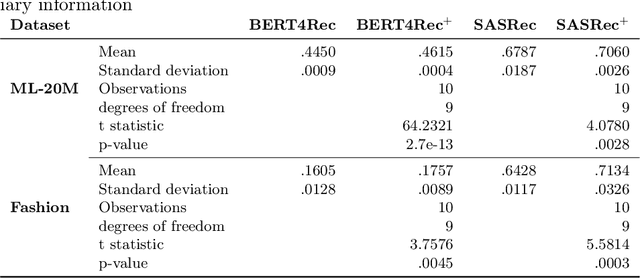
Abstract:Context-aware recommendation systems improve upon classical recommender systems by including, in the modelling, a user's behaviour. Research into context-aware recommendation systems has previously only considered the sequential ordering of items as contextual information. However, there is a wealth of unexploited additional multi-modal information available in auxiliary knowledge related to items. This study extends the existing research by evaluating a multi-modal recommendation system that exploits the inclusion of comprehensive auxiliary knowledge related to an item. The empirical results explore extracting vector representations (embeddings) from unstructured and structured data using data2vec. The fused embeddings are then used to train several state-of-the-art transformer architectures for sequential user-item representations. The analysis of the experimental results shows a statistically significant improvement in prediction accuracy, which confirms the effectiveness of including auxiliary information in a context-aware recommendation system. We report a 4% and 11% increase in the NDCG score for long and short user sequence datasets, respectively.
Dictionary learning for clustering on hyperspectral images
Feb 02, 2022Abstract:Dictionary learning and sparse coding have been widely studied as mechanisms for unsupervised feature learning. Unsupervised learning could bring enormous benefit to the processing of hyperspectral images and to other remote sensing data analysis because labelled data are often scarce in this field. We propose a method for clustering the pixels of hyperspectral images using sparse coefficients computed from a representative dictionary as features. We show empirically that the proposed method works more effectively than clustering on the original pixels. We also demonstrate that our approach, in certain circumstances, outperforms the clustering results of features extracted using principal component analysis and non-negative matrix factorisation. Furthermore, our method is suitable for applications in repetitively clustering an ever-growing amount of high-dimensional data, which is the case when working with hyperspectral satellite imagery.
* Springer Machine Learning Journal, 8 pages, 3 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge