Gareth Barker
Automated triaging of head MRI examinations using convolutional neural networks
Jun 15, 2021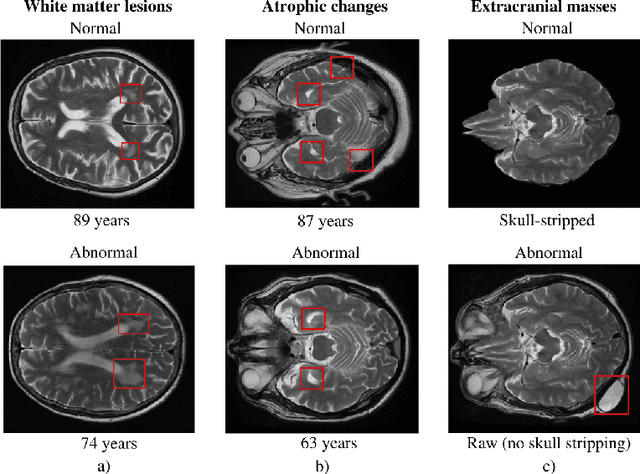
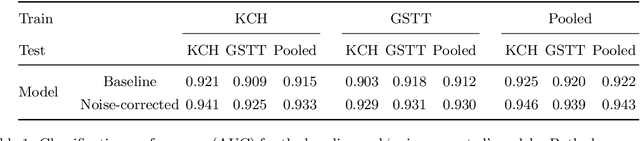
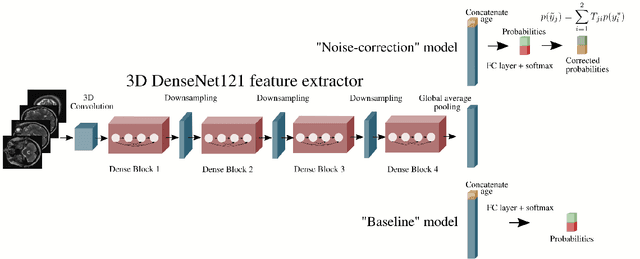
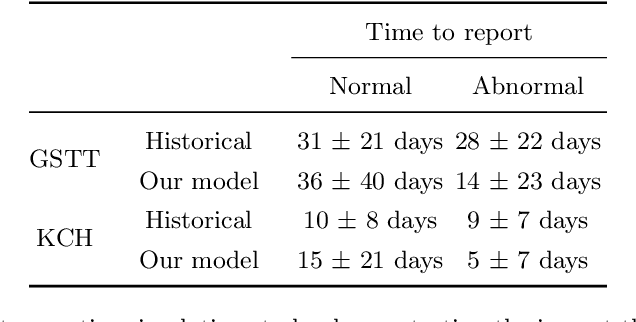
Abstract:The growing demand for head magnetic resonance imaging (MRI) examinations, along with a global shortage of radiologists, has led to an increase in the time taken to report head MRI scans around the world. For many neurological conditions, this delay can result in increased morbidity and mortality. An automated triaging tool could reduce reporting times for abnormal examinations by identifying abnormalities at the time of imaging and prioritizing the reporting of these scans. In this work, we present a convolutional neural network for detecting clinically-relevant abnormalities in $\text{T}_2$-weighted head MRI scans. Using a validated neuroradiology report classifier, we generated a labelled dataset of 43,754 scans from two large UK hospitals for model training, and demonstrate accurate classification (area under the receiver operating curve (AUC) = 0.943) on a test set of 800 scans labelled by a team of neuroradiologists. Importantly, when trained on scans from only a single hospital the model generalized to scans from the other hospital ($\Delta$AUC $\leq$ 0.02). A simulation study demonstrated that our model would reduce the mean reporting time for abnormal examinations from 28 days to 14 days and from 9 days to 5 days at the two hospitals, demonstrating feasibility for use in a clinical triage environment.
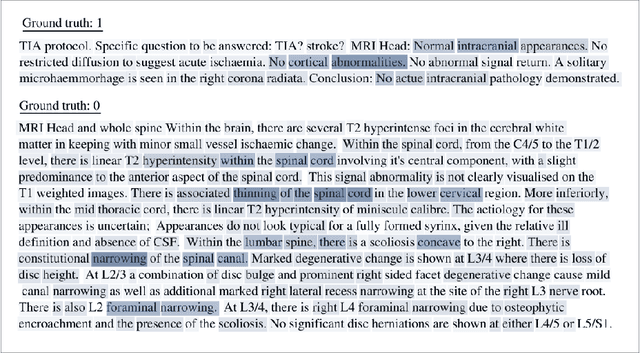
Labelling imaging datasets on the basis of neuroradiology reports: a validation study
Jul 08, 2020

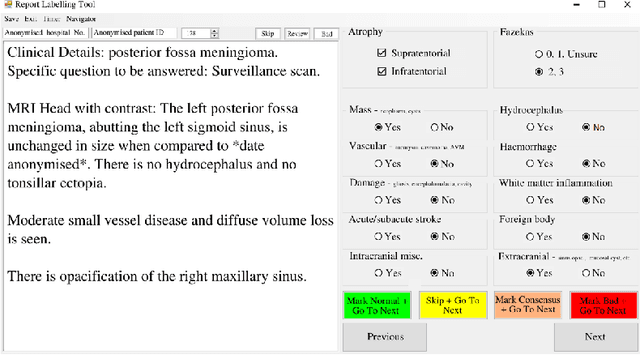
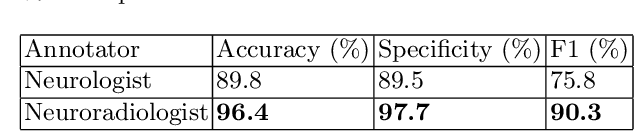
Abstract:Natural language processing (NLP) shows promise as a means to automate the labelling of hospital-scale neuroradiology magnetic resonance imaging (MRI) datasets for computer vision applications. To date, however, there has been no thorough investigation into the validity of this approach, including determining the accuracy of report labels compared to image labels as well as examining the performance of non-specialist labellers. In this work, we draw on the experience of a team of neuroradiologists who labelled over 5000 MRI neuroradiology reports as part of a project to build a dedicated deep learning-based neuroradiology report classifier. We show that, in our experience, assigning binary labels (i.e. normal vs abnormal) to images from reports alone is highly accurate. In contrast to the binary labels, however, the accuracy of more granular labelling is dependent on the category, and we highlight reasons for this discrepancy. We also show that downstream model performance is reduced when labelling of training reports is performed by a non-specialist. To allow other researchers to accelerate their research, we make our refined abnormality definitions and labelling rules available, as well as our easy-to-use radiology report labelling app which helps streamline this process.
Automated Labelling using an Attention model for Radiology reports of MRI scans (ALARM)
Feb 16, 2020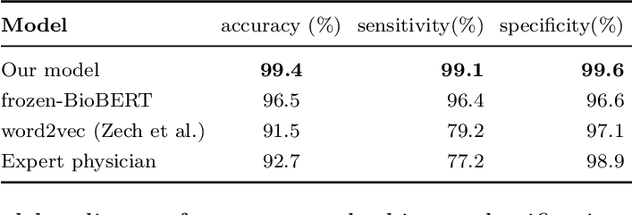
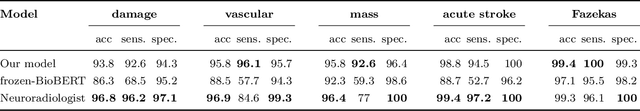
Abstract:Labelling large datasets for training high-capacity neural networks is a major obstacle to the development of deep learning-based medical imaging applications. Here we present a transformer-based network for magnetic resonance imaging (MRI) radiology report classification which automates this task by assigning image labels on the basis of free-text expert radiology reports. Our model's performance is comparable to that of an expert radiologist, and better than that of an expert physician, demonstrating the feasibility of this approach. We make code available online for researchers to label their own MRI datasets for medical imaging applications.
Nonlinear functional mapping of the human brain
Sep 08, 2015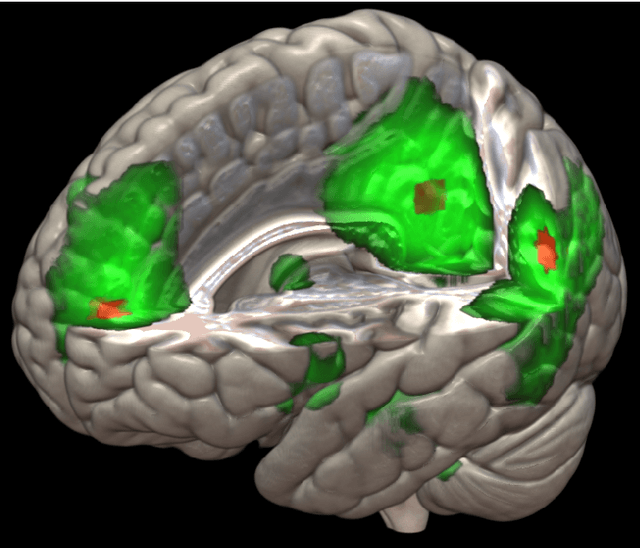
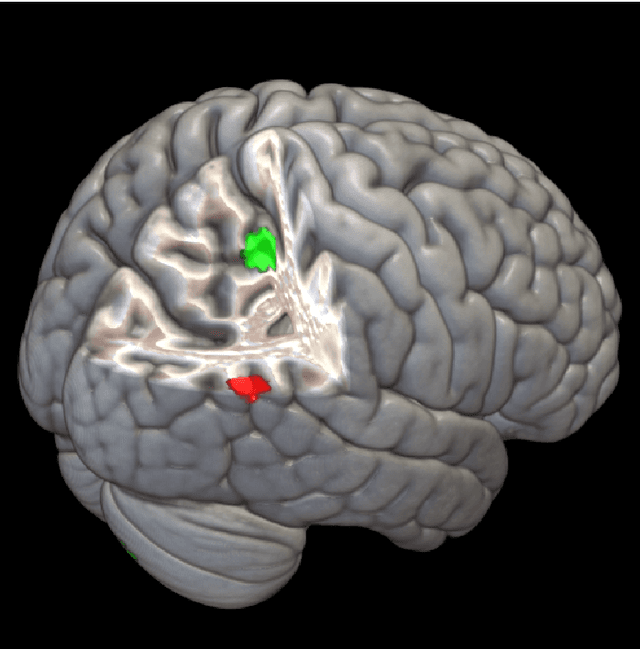
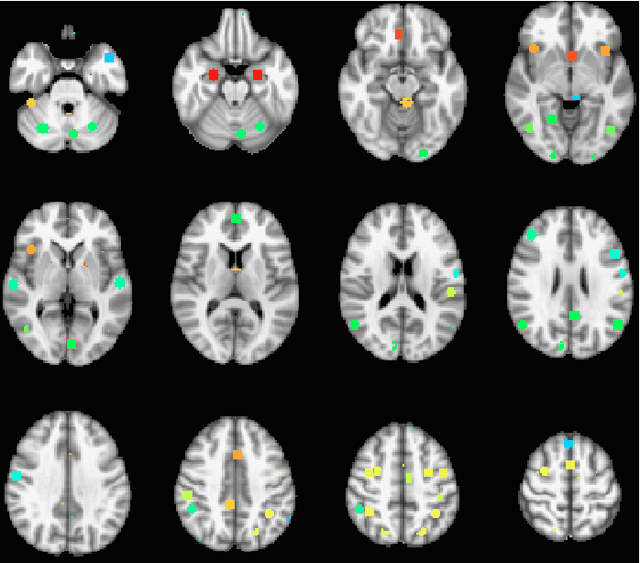
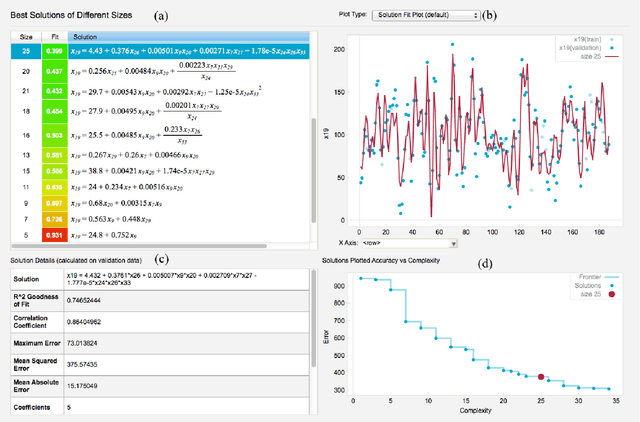
Abstract:The field of neuroimaging has truly become data rich, and novel analytical methods capable of gleaning meaningful information from large stores of imaging data are in high demand. Those methods that might also be applicable on the level of individual subjects, and thus potentially useful clinically, are of special interest. In the present study, we introduce just such a method, called nonlinear functional mapping (NFM), and demonstrate its application in the analysis of resting state fMRI from a 242-subject subset of the IMAGEN project, a European study of adolescents that includes longitudinal phenotypic, behavioral, genetic, and neuroimaging data. NFM employs a computational technique inspired by biological evolution to discover and mathematically characterize interactions among ROI (regions of interest), without making linear or univariate assumptions. We show that statistics of the resulting interaction relationships comport with recent independent work, constituting a preliminary cross-validation. Furthermore, nonlinear terms are ubiquitous in the models generated by NFM, suggesting that some of the interactions characterized here are not discoverable by standard linear methods of analysis. We discuss one such nonlinear interaction in the context of a direct comparison with a procedure involving pairwise correlation, designed to be an analogous linear version of functional mapping. We find another such interaction that suggests a novel distinction in brain function between drinking and non-drinking adolescents: a tighter coupling of ROI associated with emotion, reward, and interoceptive processes such as thirst, among drinkers. Finally, we outline many improvements and extensions of the methodology to reduce computational expense, complement other analytical tools like graph-theoretic analysis, and allow for voxel level NFM to eliminate the necessity of ROI selection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge