Fabian Bösch
Apertus: Democratizing Open and Compliant LLMs for Global Language Environments
Sep 17, 2025
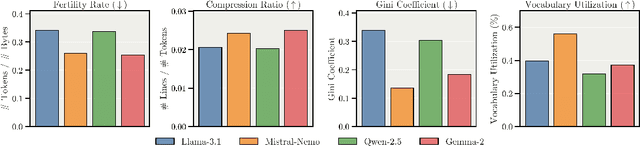

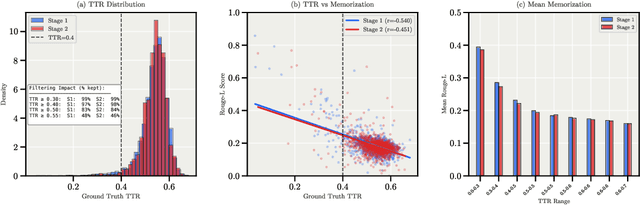
Abstract:We present Apertus, a fully open suite of large language models (LLMs) designed to address two systemic shortcomings in today's open model ecosystem: data compliance and multilingual representation. Unlike many prior models that release weights without reproducible data pipelines or regard for content-owner rights, Apertus models are pretrained exclusively on openly available data, retroactively respecting robots.txt exclusions and filtering for non-permissive, toxic, and personally identifiable content. To mitigate risks of memorization, we adopt the Goldfish objective during pretraining, strongly suppressing verbatim recall of data while retaining downstream task performance. The Apertus models also expand multilingual coverage, training on 15T tokens from over 1800 languages, with ~40% of pretraining data allocated to non-English content. Released at 8B and 70B scales, Apertus approaches state-of-the-art results among fully open models on multilingual benchmarks, rivalling or surpassing open-weight counterparts. Beyond model weights, we release all scientific artifacts from our development cycle with a permissive license, including data preparation scripts, checkpoints, evaluation suites, and training code, enabling transparent audit and extension.
Plastic Arbor: a modern simulation framework for synaptic plasticity $\unicode{x2013}$ from single synapses to networks of morphological neurons
Nov 25, 2024



Abstract:Arbor is a software library designed for efficient simulation of large-scale networks of biological neurons with detailed morphological structures. It combines customizable neuronal and synaptic mechanisms with high-performance computing, supporting multi-core CPU and GPU systems. In humans and other animals, synaptic plasticity processes play a vital role in cognitive functions, including learning and memory. Recent studies have shown that intracellular molecular processes in dendrites significantly influence single-neuron dynamics. However, for understanding how the complex interplay between dendrites and synaptic processes influences network dynamics, computational modeling is required. To enable the modeling of large-scale networks of morphologically detailed neurons with diverse plasticity processes, we have extended the Arbor library to the Plastic Arbor framework, supporting simulations of a large variety of spike-driven plasticity paradigms. To showcase the features of the new framework, we present examples of computational models, beginning with single-synapse dynamics, progressing to multi-synapse rules, and finally scaling up to large recurrent networks. While cross-validating our implementations by comparison with other simulators, we show that Arbor allows simulating plastic networks of multi-compartment neurons at nearly no additional cost in runtime compared to point-neuron simulations. Using the new framework, we have already been able to investigate the impact of dendritic structures on network dynamics across a timescale of several hours, showing a relation between the length of dendritic trees and the ability of the network to efficiently store information. By our extension of Arbor, we aim to provide a valuable tool that will support future studies on the impact of synaptic plasticity, especially, in conjunction with neuronal morphology, in large networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge