Eunha Lee
ATTNSOM: Learning Cross-Isoform Attention for Cytochrome P450 Site-of-Metabolism
Jan 28, 2026Abstract:Identifying metabolic sites where cytochrome P450 enzymes metabolize small-molecule drugs is essential for drug discovery. Although existing computational approaches have been proposed for site-of-metabolism prediction, they typically ignore cytochrome P450 isoform identity or model isoforms independently, thereby failing to fully capture inherent cross-isoform metabolic patterns. In addition, prior evaluations often rely on top-k metrics, where false positive atoms may be included among the top predictions, underscoring the need for complementary metrics that more directly assess binary atom-level discrimination under severe class imbalance. We propose ATTNSOM, an atom-level site-of-metabolism prediction framework that integrates intrinsic molecular reactivity with cross-isoform relationships. The model combines a shared graph encoder, molecule-conditioned atom representations, and a cross-attention mechanism to capture correlated metabolic patterns across cytochrome P450 isoforms. The model is evaluated on two benchmark datasets annotated with site-of-metabolism labels at atom resolution. Across these benchmarks, the model achieves consistently strong top-k performance across multiple cytochrome P450 isoforms. Relative to ablated variants, the model yields higher Matthews correlation coefficient, indicating improved discrimination of true metabolic sites. These results support the importance of explicitly modeling cross-isoform relationships for site-of-metabolism prediction. The code and datasets are available at https://github.com/dmis-lab/ATTNSOM.
Boosting CNN beyond Label in Inverse Problems
Jun 18, 2019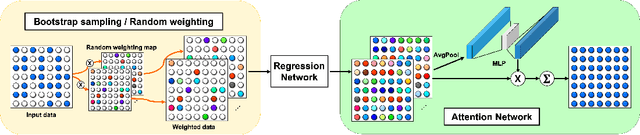

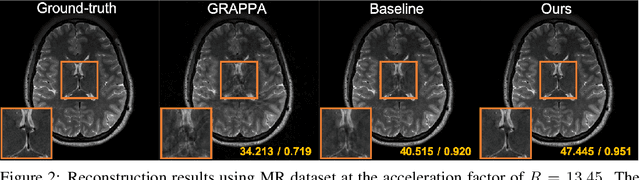
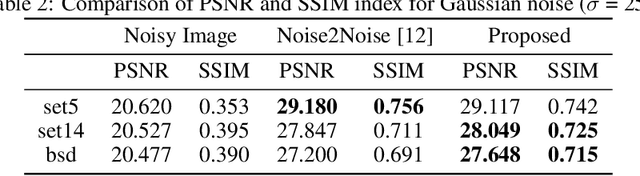
Abstract:Convolutional neural networks (CNN) have been extensively used for inverse problems. However, their prediction error for unseen test data is difficult to estimate a priori since the neural networks are trained using only selected data and their architecture are largely considered a blackbox. This poses a fundamental challenge to neural networks for unsupervised learning or improvement beyond the label. In this paper, we show that the recent unsupervised learning methods such as Noise2Noise, Stein's unbiased risk estimator (SURE)-based denoiser, and Noise2Void are closely related to each other in their formulation of an unbiased estimator of the prediction error, but each of them are associated with its own limitations. Based on these observations, we provide a novel boosting estimator for the prediction error. In particular, by employing combinatorial convolutional frame representation of encoder-decoder CNN and synergistically combining it with the batch normalization, we provide a close form formulation for the unbiased estimator of the prediction error that can be minimized for neural network training beyond the label. Experimental results show that the resulting algorithm, what we call Noise2Boosting, provides consistent improvement in various inverse problems under both supervised and unsupervised learning setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge