Dmitri B. Chklovskii
Self-calibrating Neural Networks for Dimensionality Reduction
Dec 11, 2016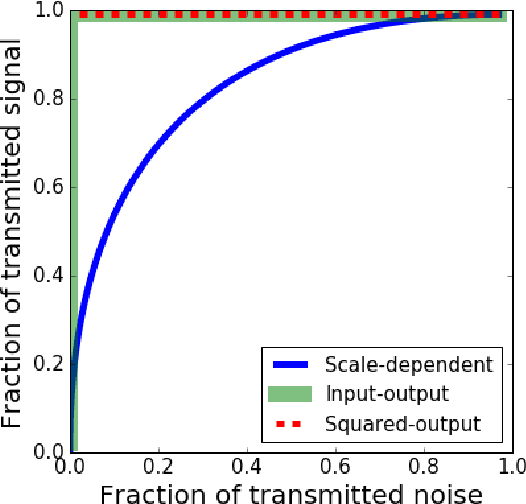
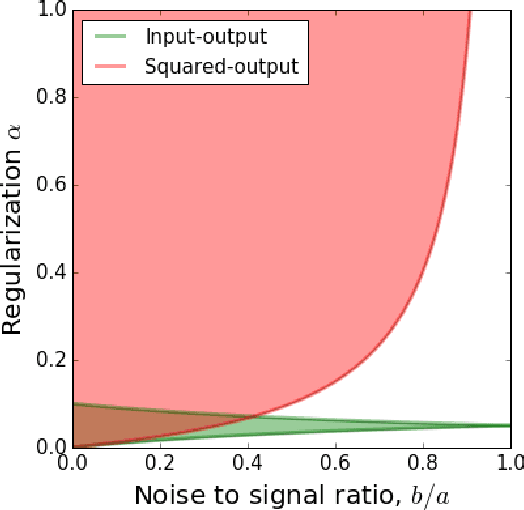
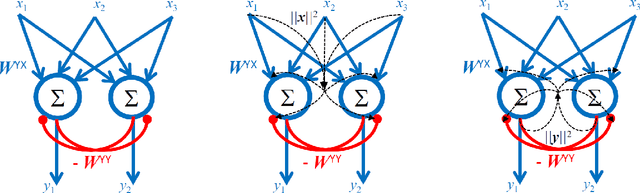
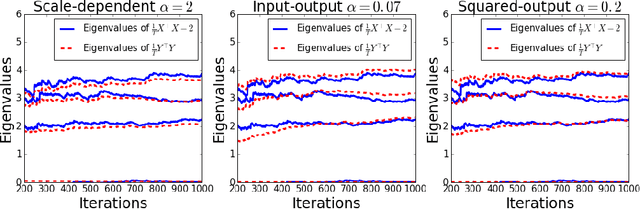
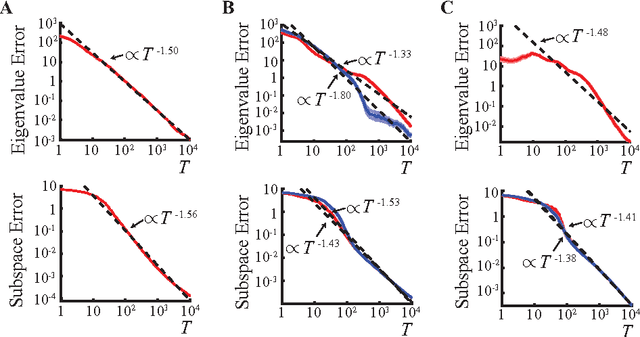
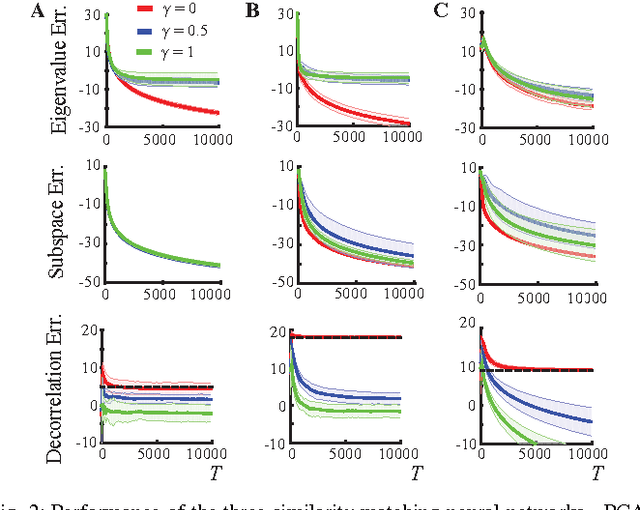
Abstract:Recently, a novel family of biologically plausible online algorithms for reducing the dimensionality of streaming data has been derived from the similarity matching principle. In these algorithms, the number of output dimensions can be determined adaptively by thresholding the singular values of the input data matrix. However, setting such threshold requires knowing the magnitude of the desired singular values in advance. Here we propose online algorithms where the threshold is self-calibrating based on the singular values computed from the existing observations. To derive these algorithms from the similarity matching cost function we propose novel regularizers. As before, these online algorithms can be implemented by Hebbian/anti-Hebbian neural networks in which the learning rule depends on the chosen regularizer. We demonstrate both mathematically and via simulation the effectiveness of these online algorithms in various settings.
A Normative Theory of Adaptive Dimensionality Reduction in Neural Networks
Jan 26, 2016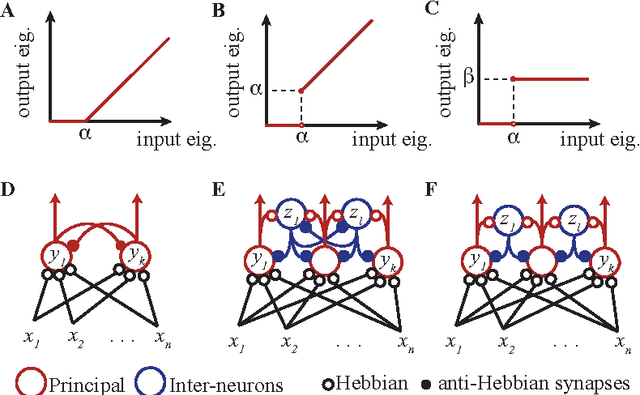
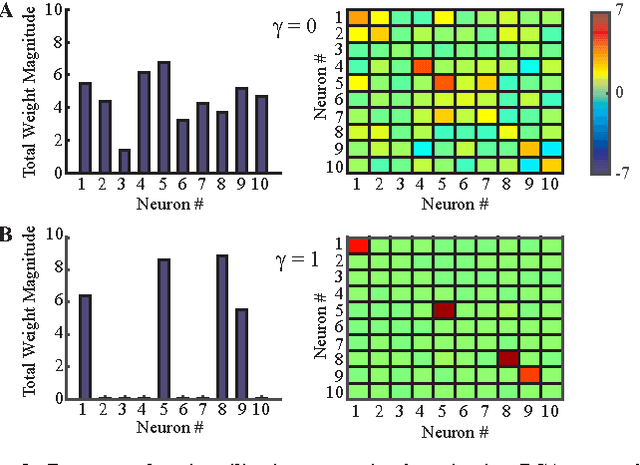
Abstract:To make sense of the world our brains must analyze high-dimensional datasets streamed by our sensory organs. Because such analysis begins with dimensionality reduction, modelling early sensory processing requires biologically plausible online dimensionality reduction algorithms. Recently, we derived such an algorithm, termed similarity matching, from a Multidimensional Scaling (MDS) objective function. However, in the existing algorithm, the number of output dimensions is set a priori by the number of output neurons and cannot be changed. Because the number of informative dimensions in sensory inputs is variable there is a need for adaptive dimensionality reduction. Here, we derive biologically plausible dimensionality reduction algorithms which adapt the number of output dimensions to the eigenspectrum of the input covariance matrix. We formulate three objective functions which, in the offline setting, are optimized by the projections of the input dataset onto its principal subspace scaled by the eigenvalues of the output covariance matrix. In turn, the output eigenvalues are computed as i) soft-thresholded, ii) hard-thresholded, iii) equalized thresholded eigenvalues of the input covariance matrix. In the online setting, we derive the three corresponding adaptive algorithms and map them onto the dynamics of neuronal activity in networks with biologically plausible local learning rules. Remarkably, in the last two networks, neurons are divided into two classes which we identify with principal neurons and interneurons in biological circuits.
Optimization theory of Hebbian/anti-Hebbian networks for PCA and whitening
Nov 30, 2015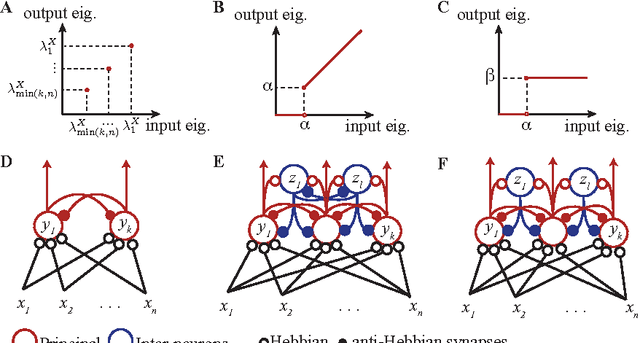
Abstract:In analyzing information streamed by sensory organs, our brains face challenges similar to those solved in statistical signal processing. This suggests that biologically plausible implementations of online signal processing algorithms may model neural computation. Here, we focus on such workhorses of signal processing as Principal Component Analysis (PCA) and whitening which maximize information transmission in the presence of noise. We adopt the similarity matching framework, recently developed for principal subspace extraction, but modify the existing objective functions by adding a decorrelating term. From the modified objective functions, we derive online PCA and whitening algorithms which are implementable by neural networks with local learning rules, i.e. synaptic weight updates that depend on the activity of only pre- and postsynaptic neurons. Our theory offers a principled model of neural computations and makes testable predictions such as the dropout of underutilized neurons.
A Hebbian/Anti-Hebbian Network for Online Sparse Dictionary Learning Derived from Symmetric Matrix Factorization
Nov 30, 2015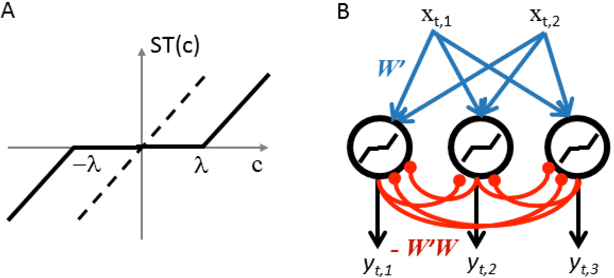
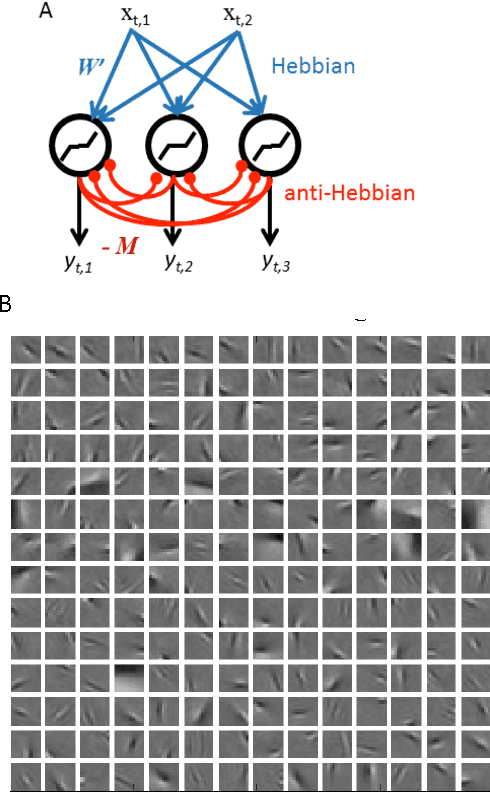
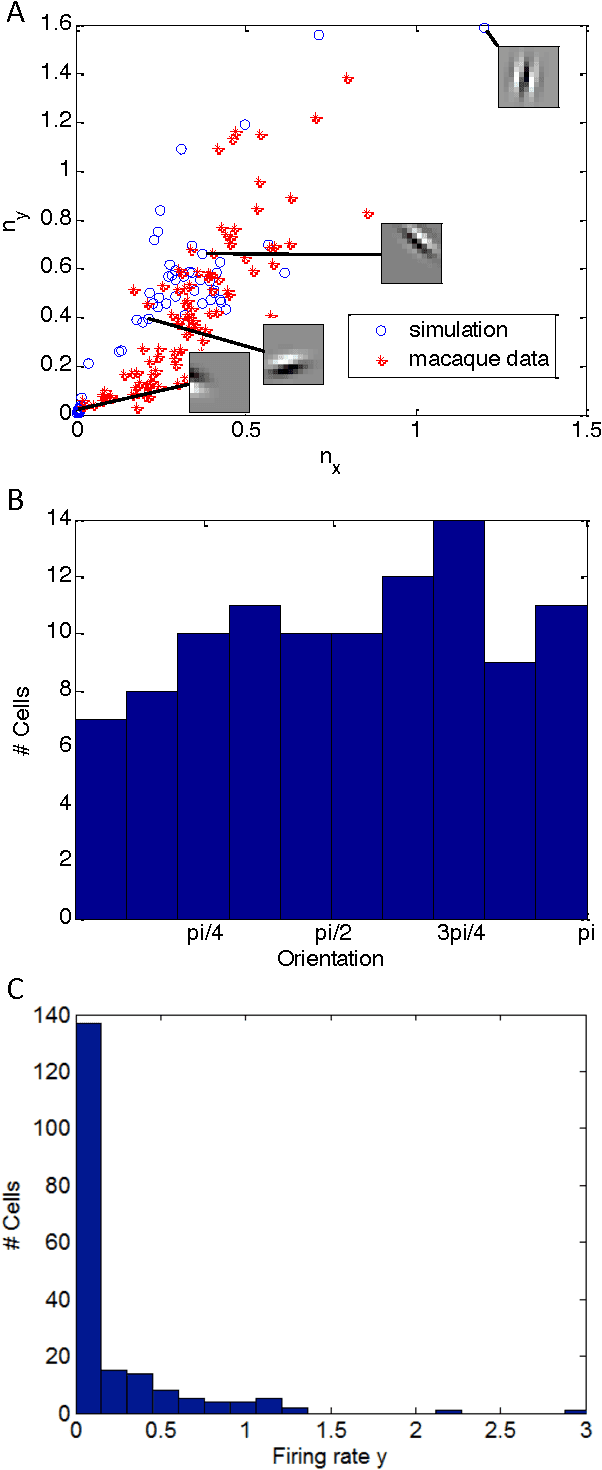
Abstract:Olshausen and Field (OF) proposed that neural computations in the primary visual cortex (V1) can be partially modeled by sparse dictionary learning. By minimizing the regularized representation error they derived an online algorithm, which learns Gabor-filter receptive fields from a natural image ensemble in agreement with physiological experiments. Whereas the OF algorithm can be mapped onto the dynamics and synaptic plasticity in a single-layer neural network, the derived learning rule is nonlocal - the synaptic weight update depends on the activity of neurons other than just pre- and postsynaptic ones - and hence biologically implausible. Here, to overcome this problem, we derive sparse dictionary learning from a novel cost-function - a regularized error of the symmetric factorization of the input's similarity matrix. Our algorithm maps onto a neural network of the same architecture as OF but using only biologically plausible local learning rules. When trained on natural images our network learns Gabor-filter receptive fields and reproduces the correlation among synaptic weights hard-wired in the OF network. Therefore, online symmetric matrix factorization may serve as an algorithmic theory of neural computation.
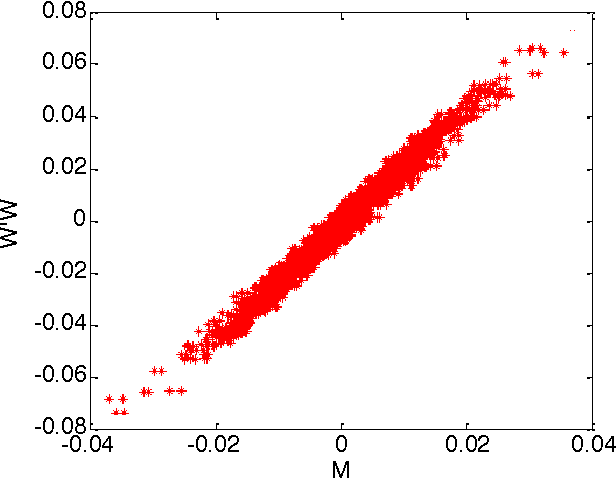
A Hebbian/Anti-Hebbian Network Derived from Online Non-Negative Matrix Factorization Can Cluster and Discover Sparse Features
Mar 02, 2015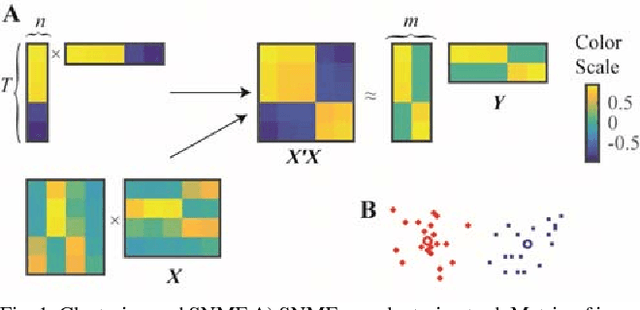
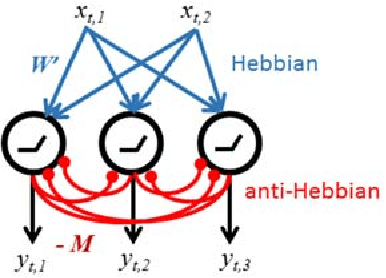
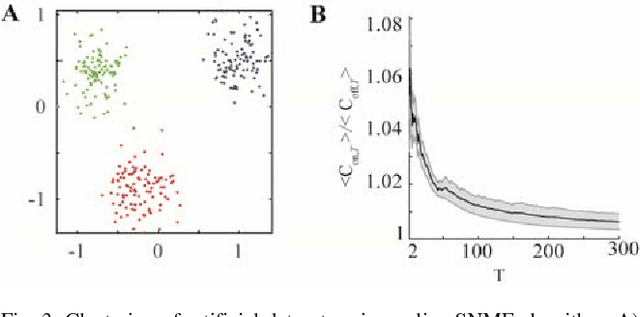
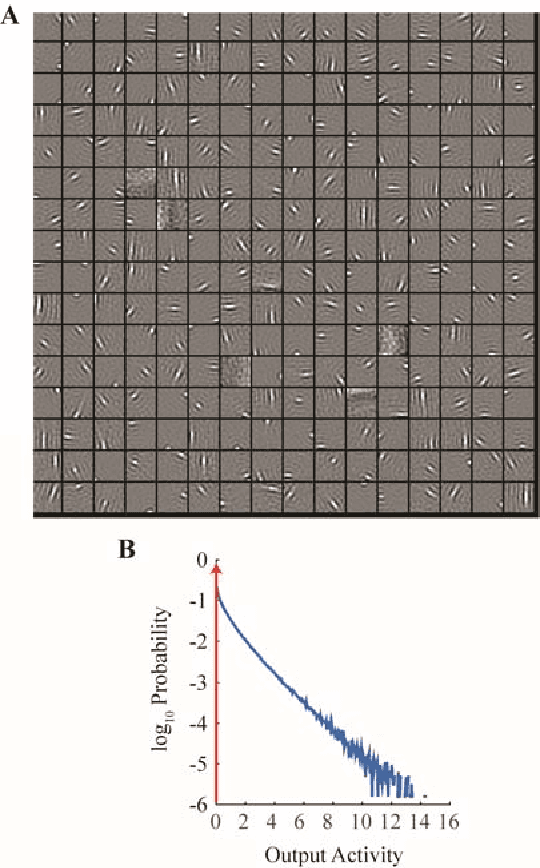
Abstract:Despite our extensive knowledge of biophysical properties of neurons, there is no commonly accepted algorithmic theory of neuronal function. Here we explore the hypothesis that single-layer neuronal networks perform online symmetric nonnegative matrix factorization (SNMF) of the similarity matrix of the streamed data. By starting with the SNMF cost function we derive an online algorithm, which can be implemented by a biologically plausible network with local learning rules. We demonstrate that such network performs soft clustering of the data as well as sparse feature discovery. The derived algorithm replicates many known aspects of sensory anatomy and biophysical properties of neurons including unipolar nature of neuronal activity and synaptic weights, local synaptic plasticity rules and the dependence of learning rate on cumulative neuronal activity. Thus, we make a step towards an algorithmic theory of neuronal function, which should facilitate large-scale neural circuit simulations and biologically inspired artificial intelligence.
A Hebbian/Anti-Hebbian Neural Network for Linear Subspace Learning: A Derivation from Multidimensional Scaling of Streaming Data
Mar 02, 2015Abstract:Neural network models of early sensory processing typically reduce the dimensionality of streaming input data. Such networks learn the principal subspace, in the sense of principal component analysis (PCA), by adjusting synaptic weights according to activity-dependent learning rules. When derived from a principled cost function these rules are nonlocal and hence biologically implausible. At the same time, biologically plausible local rules have been postulated rather than derived from a principled cost function. Here, to bridge this gap, we derive a biologically plausible network for subspace learning on streaming data by minimizing a principled cost function. In a departure from previous work, where cost was quantified by the representation, or reconstruction, error, we adopt a multidimensional scaling (MDS) cost function for streaming data. The resulting algorithm relies only on biologically plausible Hebbian and anti-Hebbian local learning rules. In a stochastic setting, synaptic weights converge to a stationary state which projects the input data onto the principal subspace. If the data are generated by a nonstationary distribution, the network can track the principal subspace. Thus, our result makes a step towards an algorithmic theory of neural computation.
A Neuron as a Signal Processing Device
May 12, 2014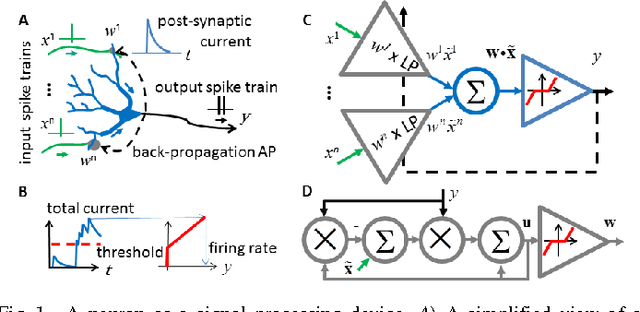
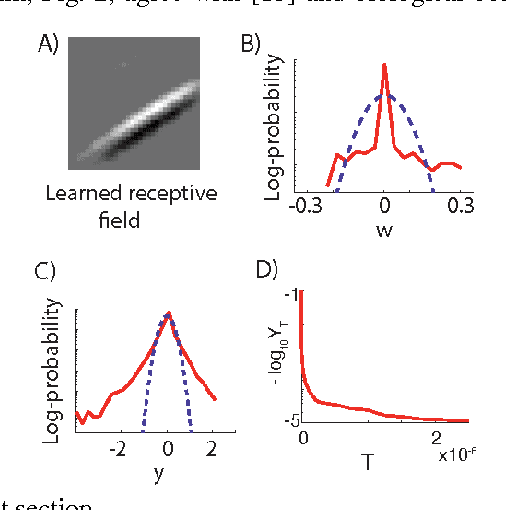
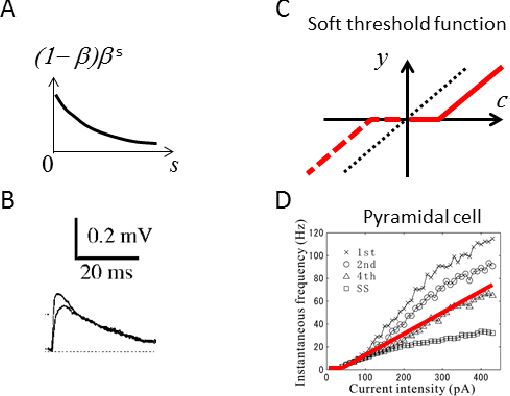
Abstract:A neuron is a basic physiological and computational unit of the brain. While much is known about the physiological properties of a neuron, its computational role is poorly understood. Here we propose to view a neuron as a signal processing device that represents the incoming streaming data matrix as a sparse vector of synaptic weights scaled by an outgoing sparse activity vector. Formally, a neuron minimizes a cost function comprising a cumulative squared representation error and regularization terms. We derive an online algorithm that minimizes such cost function by alternating between the minimization with respect to activity and with respect to synaptic weights. The steps of this algorithm reproduce well-known physiological properties of a neuron, such as weighted summation and leaky integration of synaptic inputs, as well as an Oja-like, but parameter-free, synaptic learning rule. Our theoretical framework makes several predictions, some of which can be verified by the existing data, others require further experiments. Such framework should allow modeling the function of neuronal circuits without necessarily measuring all the microscopic biophysical parameters, as well as facilitate the design of neuromorphic electronics.
Machine learning of hierarchical clustering to segment 2D and 3D images
Jul 23, 2013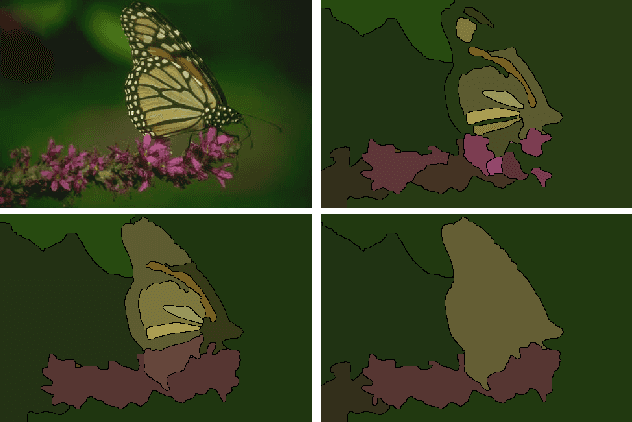
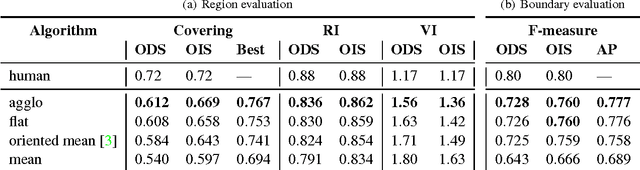
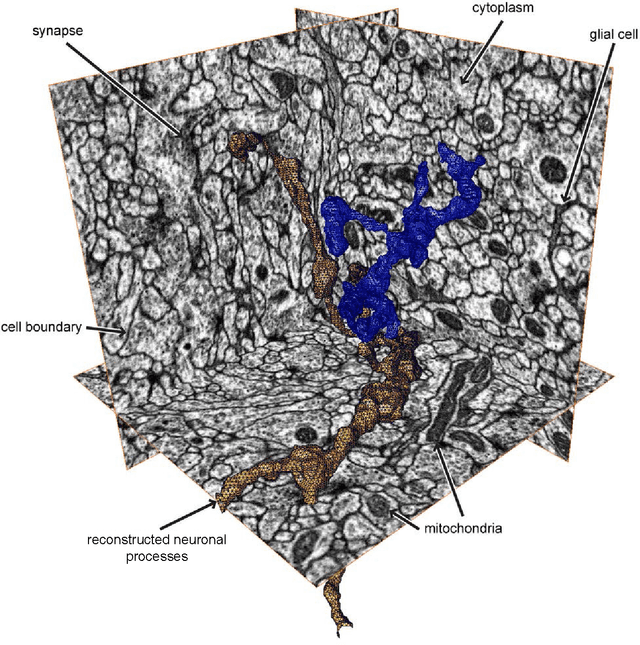
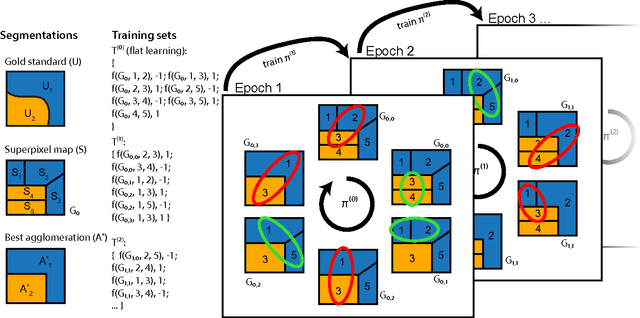
Abstract:We aim to improve segmentation through the use of machine learning tools during region agglomeration. We propose an active learning approach for performing hierarchical agglomerative segmentation from superpixels. Our method combines multiple features at all scales of the agglomerative process, works for data with an arbitrary number of dimensions, and scales to very large datasets. We advocate the use of variation of information to measure segmentation accuracy, particularly in 3D electron microscopy (EM) images of neural tissue, and using this metric demonstrate an improvement over competing algorithms in EM and natural images.
* 15 pages, 8 figures
Online computation of sparse representations of time varying stimuli using a biologically motivated neural network
Oct 13, 2012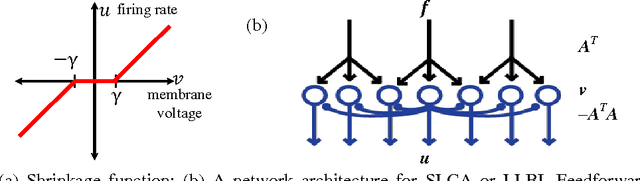
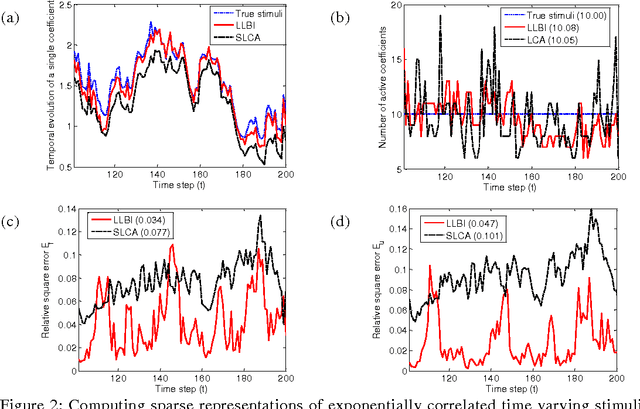
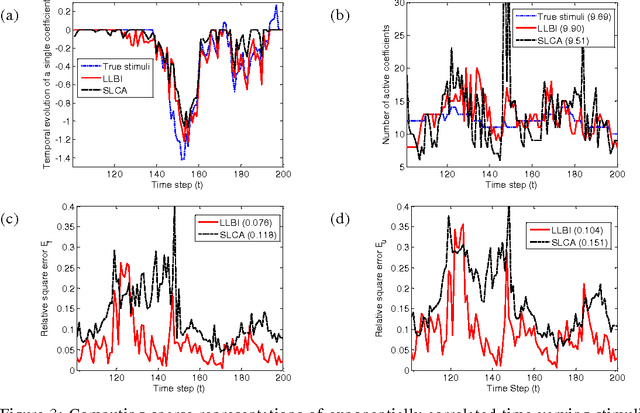
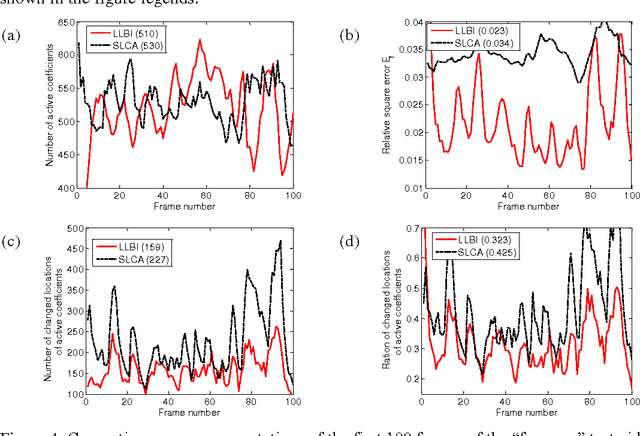
Abstract:Natural stimuli are highly redundant, possessing significant spatial and temporal correlations. While sparse coding has been proposed as an efficient strategy employed by neural systems to encode sensory stimuli, the underlying mechanisms are still not well understood. Most previous approaches model the neural dynamics by the sparse representation dictionary itself and compute the representation coefficients offline. In reality, faced with the challenge of constantly changing stimuli, neurons must compute the sparse representations dynamically in an online fashion. Here, we describe a leaky linearized Bregman iteration (LLBI) algorithm which computes the time varying sparse representations using a biologically motivated network of leaky rectifying neurons. Compared to previous attempt of dynamic sparse coding, LLBI exploits the temporal correlation of stimuli and demonstrate better performance both in representation error and the smoothness of temporal evolution of sparse coefficients.
A network of spiking neurons for computing sparse representations in an energy efficient way
Oct 04, 2012Abstract:Computing sparse redundant representations is an important problem both in applied mathematics and neuroscience. In many applications, this problem must be solved in an energy efficient way. Here, we propose a hybrid distributed algorithm (HDA), which solves this problem on a network of simple nodes communicating via low-bandwidth channels. HDA nodes perform both gradient-descent-like steps on analog internal variables and coordinate-descent-like steps via quantized external variables communicated to each other. Interestingly, such operation is equivalent to a network of integrate-and-fire neurons, suggesting that HDA may serve as a model of neural computation. We show that the numerical performance of HDA is on par with existing algorithms. In the asymptotic regime the representation error of HDA decays with time, t, as 1/t. HDA is stable against time-varying noise, specifically, the representation error decays as 1/sqrt(t) for Gaussian white noise.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge