Dimos Baltas
Analysis of clinical, dosimetric and radiomic features for predicting local failure after stereotactic radiotherapy of brain metastases in malignant melanoma
May 31, 2024



Abstract:Background: The aim of this study was to investigate the role of clinical, dosimetric and pretherapeutic magnetic resonance imaging (MRI) features for lesion-specific outcome prediction of stereotactic radiotherapy (SRT) in patients with brain metastases from malignant melanoma (MBM). Methods: In this multicenter, retrospective analysis, we reviewed 517 MBM from 130 patients treated with SRT (single fraction or hypofractionated). For each gross tumor volume (GTV) 1576 radiomic features (RF) were calculated (788 each for the GTV and for a 3 mm margin around the GTV). Clinical parameters, radiation dose and RF from pretherapeutic contrast-enhanced T1-weighted MRI from different institutions were evaluated with a feature processing and elimination pipeline in a nested cross-validation scheme. Results: Seventy-two (72) of 517 lesions (13.9%) showed a local failure (LF) after SRT. The processing pipeline showed clinical, dosimetric and radiomic features providing information for LF prediction. The most prominent ones were the correlation of the gray level co-occurrence matrix of the margin (hazard ratio (HR): 0.37, confidence interval (CI): 0.23-0.58) and systemic therapy before SRT (HR: 0.55, CI: 0.42-0.70). The majority of RF associated with LF was calculated in the margin around the GTV. Conclusions: Pretherapeutic MRI based RF connected with lesion-specific outcome after SRT could be identified, despite multicentric data and minor differences in imaging protocols. Image data analysis of the surrounding metastatic environment may provide therapy-relevant information with the potential to further individualize radiotherapy strategies.
The use of deep learning in interventional radiotherapy : a review with a focus on open source and open data
May 16, 2022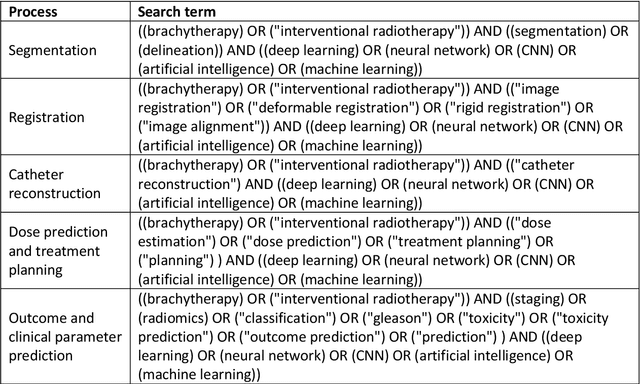
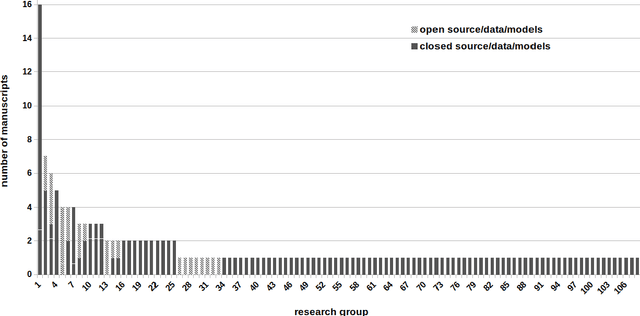
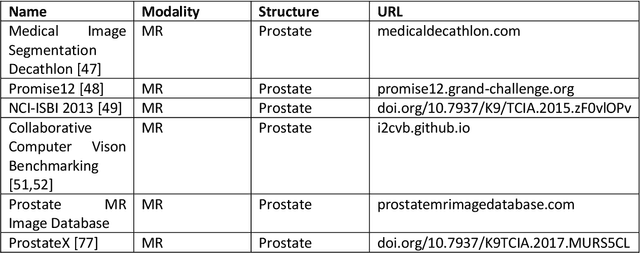
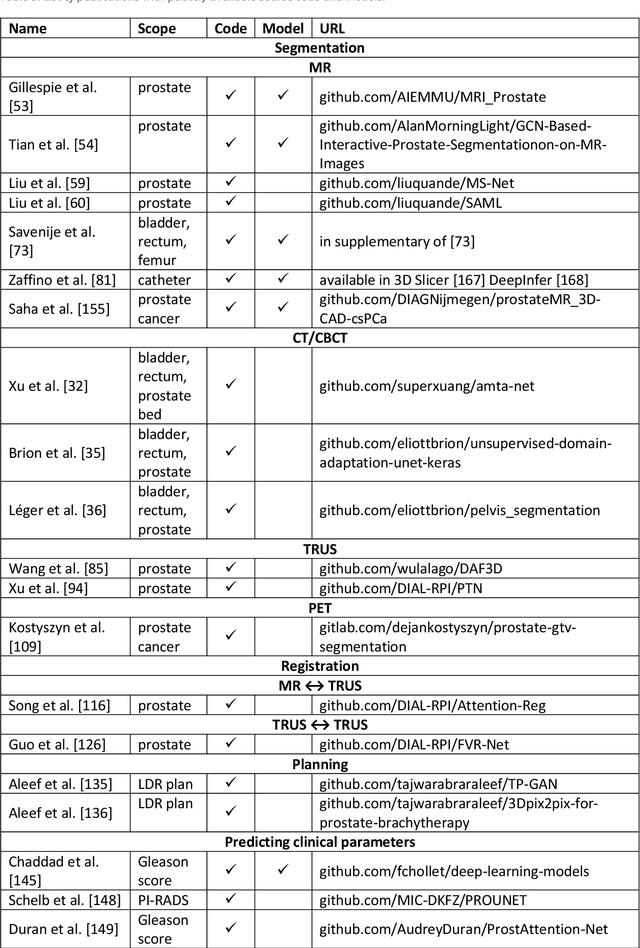
Abstract:Deep learning advanced to one of the most important technologies in almost all medical fields. Especially in areas, related to medical imaging it plays a big role. However, in interventional radiotherapy (brachytherapy) deep learning is still in an early phase. In this review, first, we investigated and scrutinised the role of deep learning in all processes of interventional radiotherapy and directly related fields. Additionally we summarised the most recent developments. To reproduce results of deep learning algorithms both source code and training data must be available. Therefore, a second focus of this work was on the analysis of the availability of open source, open data and open models. In our analysis, we were able to show that deep learning plays already a major role in some areas of interventional radiotherapy, but is still hardly presented in others. Nevertheless, its impact is increasing with the years, partly self-propelled but also influenced by closely related fields. Open source, data and models are growing in number but are still scarce and unevenly distributed among different research groups. The reluctance in publishing code, data and models limits reproducibility and restricts evaluation to mono-institutional datasets. Summarised, deep learning will change positively the workflow of interventional radiotherapy but there is room for improvement when it comes to reproducible results and standardised evaluation methods.
Measuring breathing induced oesophageal motion and its dosimetric impact
Oct 19, 2020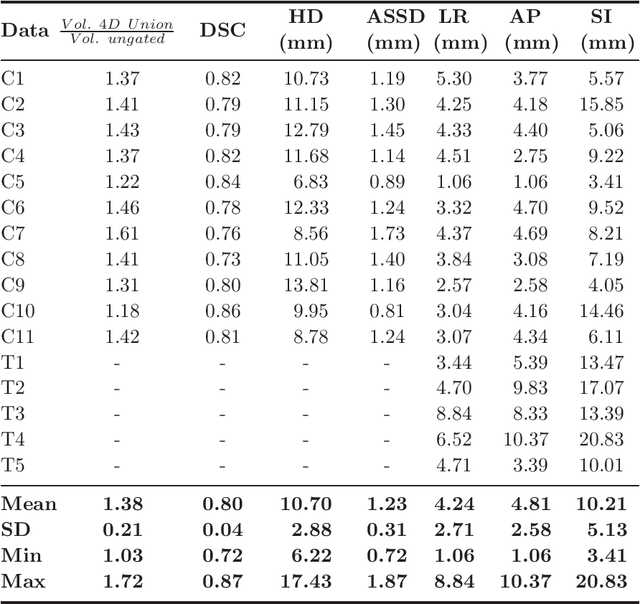
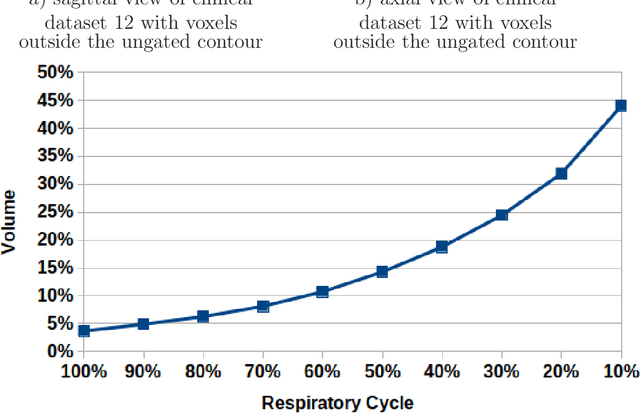
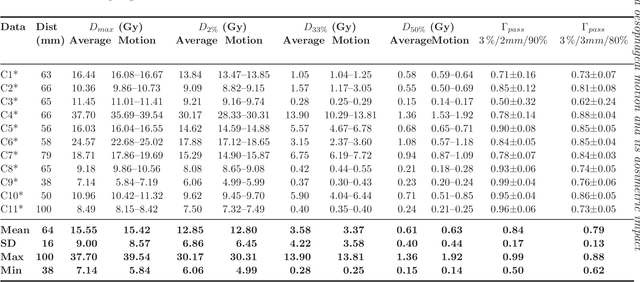
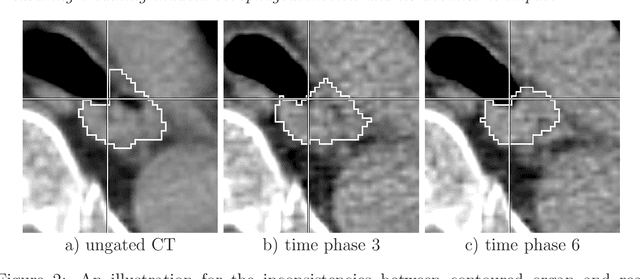
Abstract:Stereotactic body radiation therapy allows for a precise and accurate dose delivery. Organ motion during treatment bares the risk of undetected high dose healthy tissue exposure. An organ very susceptible to high dose is the oesophagus. Its low contrast on CT and the oblong shape renders motion estimation difficult. We tackle this issue by modern algorithms to measure the oesophageal motion voxel-wise and to estimate motion related dosimetric impact. Oesophageal motion was measured using deformable image registration and 4DCT of 11 internal and 5 public datasets. Current clinical practice of contouring the organ on 3DCT was compared to timely resolved 4DCT contours. The dosimetric impact of the motion was estimated by analysing the trajectory of each voxel in the 4D dose distribution. Finally an organ motion model was built, allowing for easier patient-wise comparisons. Motion analysis showed mean absolute maximal motion amplitudes of 4.24 +/- 2.71 mm left-right, 4.81 +/- 2.58 mm anterior-posterior and 10.21 +/- 5.13 mm superior-inferior. Motion between the cohorts differed significantly. In around 50 % of the cases the dosimetric passing criteria was violated. Contours created on 3DCT did not cover 14 % of the organ for 50 % of the respiratory cycle and the 3D contour is around 38 % smaller than the union of all 4D contours. The motion model revealed that the maximal motion is not limited to the lower part of the organ. Our results showed motion amplitudes higher than most reported values in the literature and that motion is very heterogeneous across patients. Therefore, individual motion information should be considered in contouring and planning.
Convolutional neural network based deep-learning architecture for intraprostatic tumour contouring on PSMA PET images in patients with primary prostate cancer
Aug 07, 2020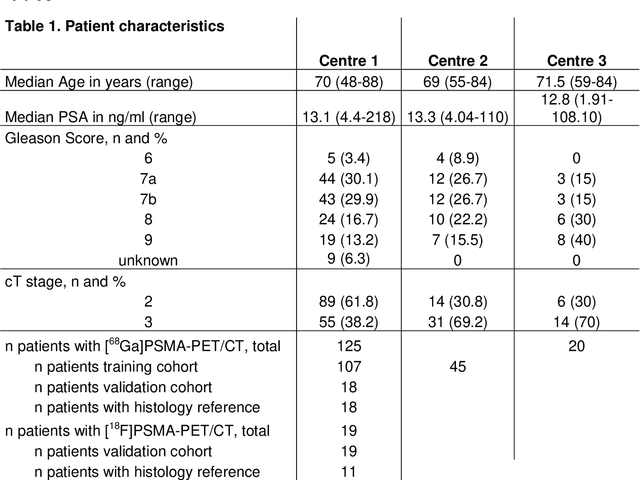
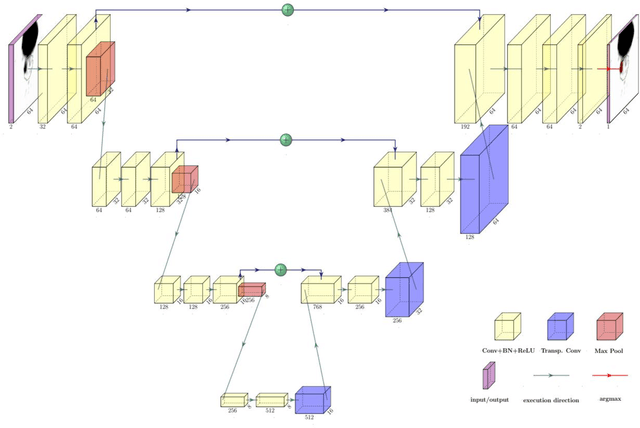
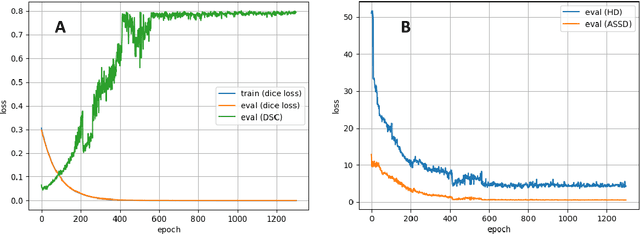
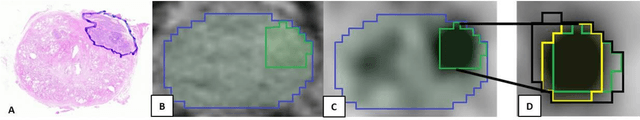
Abstract:Accurate delineation of the intraprostatic gross tumour volume (GTV) is a prerequisite for treatment approaches in patients with primary prostate cancer (PCa). Prostate-specific membrane antigen positron emission tomography (PSMA-PET) may outperform MRI in GTV detection. However, visual GTV delineation underlies interobserver heterogeneity and is time consuming. The aim of this study was to develop a convolutional neural network (CNN) for automated segmentation of intraprostatic tumour (GTV-CNN) in PSMA-PET. Methods: The CNN (3D U-Net) was trained on [68Ga]PSMA-PET images of 152 patients from two different institutions and the training labels were generated manually using a validated technique. The CNN was tested on two independent internal (cohort 1: [68Ga]PSMA-PET, n=18 and cohort 2: [18F]PSMA-PET, n=19) and one external (cohort 3: [68Ga]PSMA-PET, n=20) test-datasets. Accordance between manual contours and GTV-CNN was assessed with Dice-S{\o}rensen coefficient (DSC). Sensitivity and specificity were calculated for the two internal test-datasets by using whole-mount histology. Results: Median DSCs for cohorts 1-3 were 0.84 (range: 0.32-0.95), 0.81 (range: 0.28-0.93) and 0.83 (range: 0.32-0.93), respectively. Sensitivities and specificities for GTV-CNN were comparable with manual expert contours: 0.98 and 0.76 (cohort 1) and 1 and 0.57 (cohort 2), respectively. Computation time was around 6 seconds for a standard dataset. Conclusion: The application of a CNN for automated contouring of intraprostatic GTV in [68Ga]PSMA- and [18F]PSMA-PET images resulted in a high concordance with expert contours and in high sensitivities and specificities in comparison with histology reference. This robust, accurate and fast technique may be implemented for treatment concepts in primary PCa. The trained model and the study's source code are available in an open source repository.
One Shot Learning for Deformable Medical Image Registration and Periodic Motion Tracking
Jul 11, 2019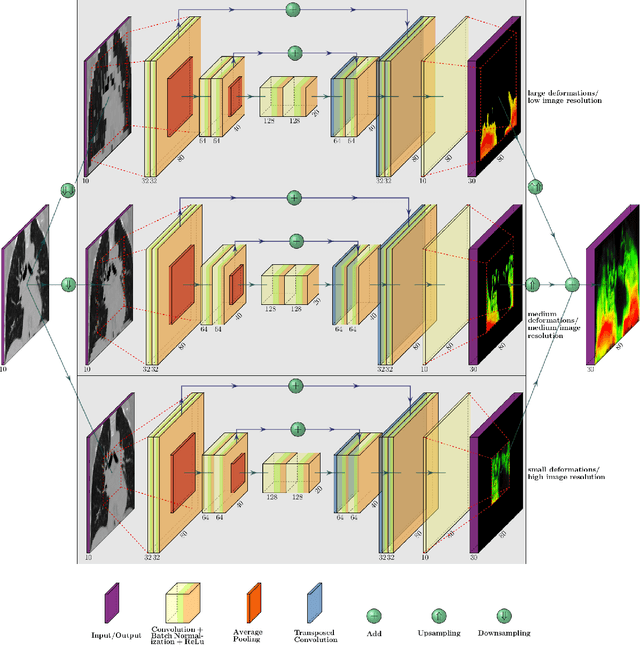
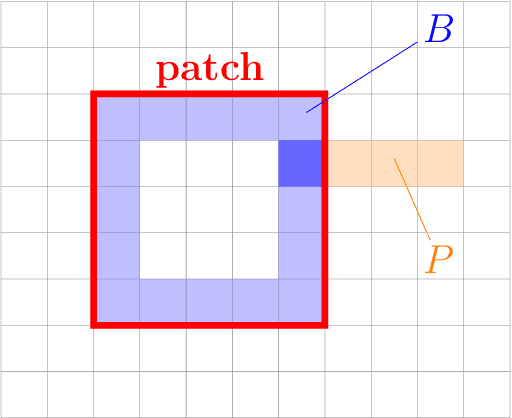
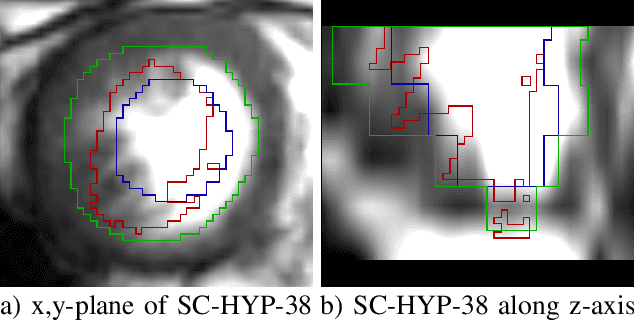
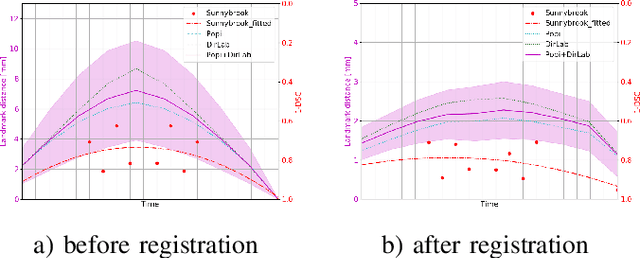
Abstract:Deformable image registration is a very important field of research in medical imaging. Recently multiple deep learning approaches were published in this area showing promising results. However, drawbacks of deep learning methods are the need for a large amount of training datasets and their inability to register unseen images different from the training datasets. One shot learning comes without the need of large training datasets and has already been proven to be applicable to 3D data. In this work we present an one shot registration approach for periodic motion tracking in 3D and 4D datasets. When applied to 3D dataset the algorithm calculates the inverse of a registration vector field simultaneously. For registration we employed a U-Net combined with a coarse to fine approach and a differential spatial transformer module. The algorithm was thoroughly tested with multiple 4D and 3D datasets publicly available. The results show that the presented approach is able to track periodic motion and to yield a competitive registration accuracy. Possible applications are the use as a stand-alone algorithm for 3D and 4D motion tracking or in the beginning of studies until enough datasets for a separate training phase are available.
A 3D fully convolutional neural network and a random walker to segment the esophagus in CT
Apr 21, 2017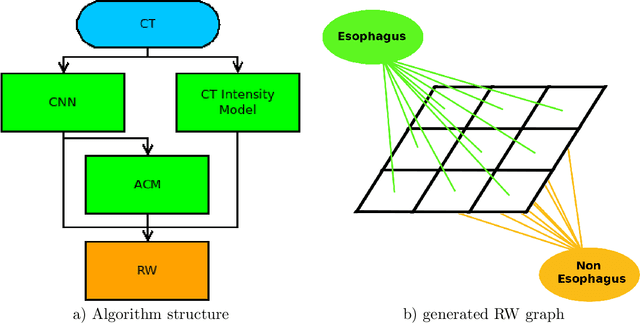
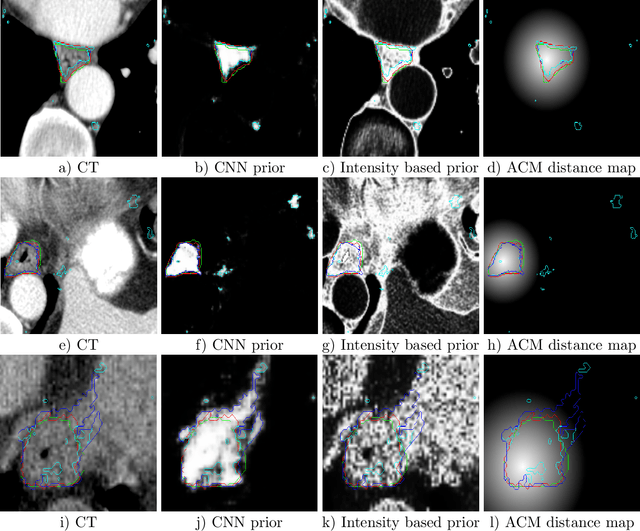
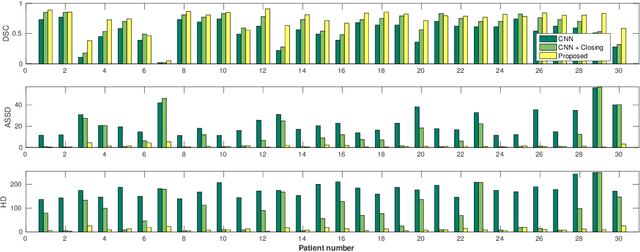
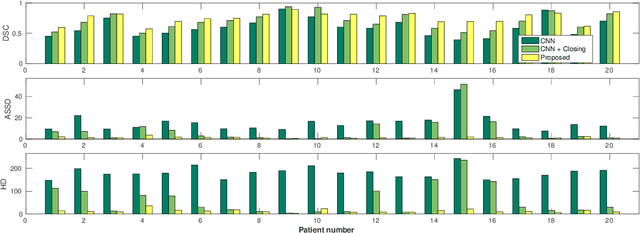
Abstract:Precise delineation of organs at risk (OAR) is a crucial task in radiotherapy treatment planning, which aims at delivering high dose to the tumour while sparing healthy tissues. In recent years algorithms showed high performance and the possibility to automate this task for many OAR. However, for some OAR precise delineation remains challenging. The esophagus with a versatile shape and poor contrast is among these structures. To tackle these issues we propose a 3D fully (convolutional neural network (CNN) driven random walk (RW) approach to automatically segment the esophagus on CT. First, a soft probability map is generated by the CNN. Then an active contour model (ACM) is fitted on the probability map to get a first estimation of the center line. The outputs of the CNN and ACM are then used in addition to CT Hounsfield values to drive the RW. Evaluation and training was done on 50 CTs with peer reviewed esophagus contours. Results were assessed regarding spatial overlap and shape similarities. The generated contours showed a mean Dice coefficient of 0.76, an average symmetric square distance of 1.36 mm and an average Hausdorff distance of 11.68 compared to the reference. These figures translate into a very good agreement with the reference contours and an increase in accuracy compared to other methods. We show that by employing a CNN accurate estimations of esophagus location can be obtained and refined by a post processing RW step. One of the main advantages compared to previous methods is that our network performs convolutions in a 3D manner, fully exploiting the 3D spatial context and performing an efficient and precise volume-wise prediction. The whole segmentation process is fully automatic and yields esophagus delineations in very good agreement with the used gold standard, showing that it can compete with previously published methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge