Dimitris Spathis
FairComp: Workshop on Fairness and Robustness in Machine Learning for Ubiquitous Computing
Sep 22, 2023Abstract:How can we ensure that Ubiquitous Computing (UbiComp) research outcomes are both ethical and fair? While fairness in machine learning (ML) has gained traction in recent years, fairness in UbiComp remains unexplored. This workshop aims to discuss fairness in UbiComp research and its social, technical, and legal implications. From a social perspective, we will examine the relationship between fairness and UbiComp research and identify pathways to ensure that ubiquitous technologies do not cause harm or infringe on individual rights. From a technical perspective, we will initiate a discussion on data practices to develop bias mitigation approaches tailored to UbiComp research. From a legal perspective, we will examine how new policies shape our community's work and future research. We aim to foster a vibrant community centered around the topic of responsible UbiComp, while also charting a clear path for future research endeavours in this field.
The first step is the hardest: Pitfalls of Representing and Tokenizing Temporal Data for Large Language Models
Sep 12, 2023
Abstract:Large Language Models (LLMs) have demonstrated remarkable generalization across diverse tasks, leading individuals to increasingly use them as personal assistants and universal computing engines. Nevertheless, a notable obstacle emerges when feeding numerical/temporal data into these models, such as data sourced from wearables or electronic health records. LLMs employ tokenizers in their input that break down text into smaller units. However, tokenizers are not designed to represent numerical values and might struggle to understand repetitive patterns and context, treating consecutive values as separate tokens and disregarding their temporal relationships. Here, we discuss recent works that employ LLMs for human-centric tasks such as in mobile health sensing and present a case study showing that popular LLMs tokenize temporal data incorrectly. To address that, we highlight potential solutions such as prompt tuning with lightweight embedding layers as well as multimodal adapters, that can help bridge this "modality gap". While the capability of language models to generalize to other modalities with minimal or no finetuning is exciting, this paper underscores the fact that their outputs cannot be meaningful if they stumble over input nuances.
UDAMA: Unsupervised Domain Adaptation through Multi-discriminator Adversarial Training with Noisy Labels Improves Cardio-fitness Prediction
Jul 31, 2023



Abstract:Deep learning models have shown great promise in various healthcare monitoring applications. However, most healthcare datasets with high-quality (gold-standard) labels are small-scale, as directly collecting ground truth is often costly and time-consuming. As a result, models developed and validated on small-scale datasets often suffer from overfitting and do not generalize well to unseen scenarios. At the same time, large amounts of imprecise (silver-standard) labeled data, annotated by approximate methods with the help of modern wearables and in the absence of ground truth validation, are starting to emerge. However, due to measurement differences, this data displays significant label distribution shifts, which motivates the use of domain adaptation. To this end, we introduce UDAMA, a method with two key components: Unsupervised Domain Adaptation and Multidiscriminator Adversarial Training, where we pre-train on the silver-standard data and employ adversarial adaptation with the gold-standard data along with two domain discriminators. In particular, we showcase the practical potential of UDAMA by applying it to Cardio-respiratory fitness (CRF) prediction. CRF is a crucial determinant of metabolic disease and mortality, and it presents labels with various levels of noise (goldand silver-standard), making it challenging to establish an accurate prediction model. Our results show promising performance by alleviating distribution shifts in various label shift settings. Additionally, by using data from two free-living cohort studies (Fenland and BBVS), we show that UDAMA consistently outperforms up to 12% compared to competitive transfer learning and state-of-the-art domain adaptation models, paving the way for leveraging noisy labeled data to improve fitness estimation at scale.
Latent Masking for Multimodal Self-supervised Learning in Health Timeseries
Jul 31, 2023Abstract:Limited availability of labeled data for machine learning on biomedical time-series hampers progress in the field. Self-supervised learning (SSL) is a promising approach to learning data representations without labels. However, current SSL methods require expensive computations for negative pairs and are designed for single modalities, limiting their versatility. To overcome these limitations, we introduce CroSSL (Cross-modal SSL). CroSSL introduces two novel concepts: masking intermediate embeddings from modality-specific encoders and aggregating them into a global embedding using a cross-modal aggregator. This enables the handling of missing modalities and end-to-end learning of cross-modal patterns without prior data preprocessing or time-consuming negative-pair sampling. We evaluate CroSSL on various multimodal time-series benchmarks, including both medical-grade and consumer biosignals. Our results demonstrate superior performance compared to previous SSL techniques and supervised benchmarks with minimal labeled data. We additionally analyze the impact of different masking ratios and strategies and assess the robustness of the learned representations to missing modalities. Overall, our work achieves state-of-the-art performance while highlighting the benefits of masking latent embeddings for cross-modal learning in temporal health data.
Practical self-supervised continual learning with continual fine-tuning
Mar 30, 2023



Abstract:Self-supervised learning (SSL) has shown remarkable performance in computer vision tasks when trained offline. However, in a Continual Learning (CL) scenario where new data is introduced progressively, models still suffer from catastrophic forgetting. Retraining a model from scratch to adapt to newly generated data is time-consuming and inefficient. Previous approaches suggested re-purposing self-supervised objectives with knowledge distillation to mitigate forgetting across tasks, assuming that labels from all tasks are available during fine-tuning. In this paper, we generalize self-supervised continual learning in a practical setting where available labels can be leveraged in any step of the SSL process. With an increasing number of continual tasks, this offers more flexibility in the pre-training and fine-tuning phases. With Kaizen, we introduce a training architecture that is able to mitigate catastrophic forgetting for both the feature extractor and classifier with a carefully designed loss function. By using a set of comprehensive evaluation metrics reflecting different aspects of continual learning, we demonstrated that Kaizen significantly outperforms previous SSL models in competitive vision benchmarks, with up to 16.5% accuracy improvement on split CIFAR-100. Kaizen is able to balance the trade-off between knowledge retention and learning from new data with an end-to-end model, paving the way for practical deployment of continual learning systems.
Beyond Accuracy: A Critical Review of Fairness in Machine Learning for Mobile and Wearable Computing
Mar 27, 2023



Abstract:The field of mobile, wearable, and ubiquitous computing (UbiComp) is undergoing a revolutionary integration of machine learning. Devices can now diagnose diseases, predict heart irregularities, and unlock the full potential of human cognition. However, the underlying algorithms are not immune to biases with respect to sensitive attributes (e.g., gender, race), leading to discriminatory outcomes. The research communities of HCI and AI-Ethics have recently started to explore ways of reporting information about datasets to surface and, eventually, counter those biases. The goal of this work is to explore the extent to which the UbiComp community has adopted such ways of reporting and highlight potential shortcomings. Through a systematic review of papers published in the Proceedings of the ACM Interactive, Mobile, Wearable and Ubiquitous Technologies (IMWUT) journal over the past 5 years (2018-2022), we found that progress on algorithmic fairness within the UbiComp community lags behind. Our findings show that only a small portion (5%) of published papers adheres to modern fairness reporting, while the overwhelming majority thereof focuses on accuracy or error metrics. In light of these findings, our work provides practical guidelines for the design and development of ubiquitous technologies that not only strive for accuracy but also for fairness.
Turning Silver into Gold: Domain Adaptation with Noisy Labels for Wearable Cardio-Respiratory Fitness Prediction
Nov 20, 2022Abstract:Deep learning models have shown great promise in various healthcare applications. However, most models are developed and validated on small-scale datasets, as collecting high-quality (gold-standard) labels for health applications is often costly and time-consuming. As a result, these models may suffer from overfitting and not generalize well to unseen data. At the same time, an extensive amount of data with imprecise labels (silver-standard) is starting to be generally available, as collected from inexpensive wearables like accelerometers and electrocardiography sensors. These currently underutilized datasets and labels can be leveraged to produce more accurate clinical models. In this work, we propose UDAMA, a novel model with two key components: Unsupervised Domain Adaptation and Multi-discriminator Adversarial training, which leverage noisy data from source domain (the silver-standard dataset) to improve gold-standard modeling. We validate our framework on the challenging task of predicting lab-measured maximal oxygen consumption (VO$_{2}$max), the benchmark metric of cardio-respiratory fitness, using free-living wearable sensor data from two cohort studies as inputs. Our experiments show that the proposed framework achieves the best performance of corr = 0.665 $\pm$ 0.04, paving the way for accurate fitness estimation at scale.
Looking for Out-of-Distribution Environments in Critical Care: A case study with the eICU Database
May 26, 2022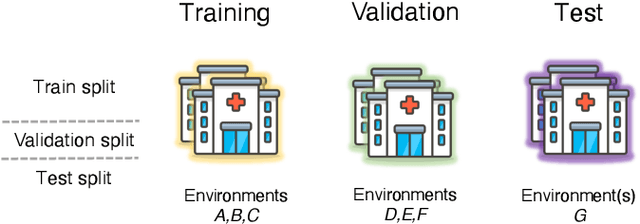
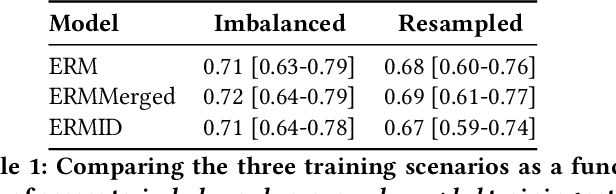
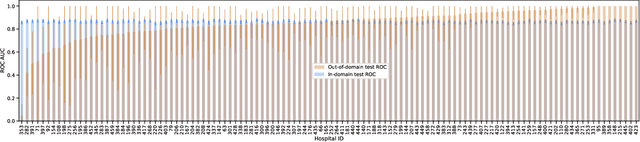
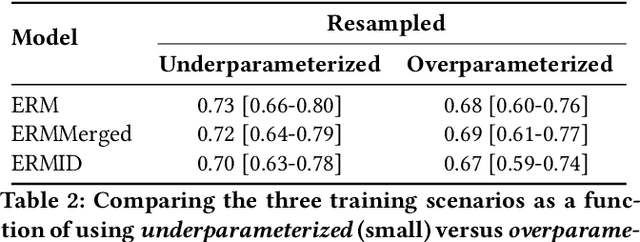
Abstract:Generalizing to new populations and domains in machine learning is still an open problem which has seen increased interest recently. In particular, clinical models show a significant performance drop when tested in settings not seen during training, e.g., new hospitals or population demographics. Recently proposed models for domain generalisation promise to alleviate this problem by learning invariant characteristics across environments, however, there is still scepticism about whether they improve over traditional training. In this work, we take a principled approach to identifying Out of Distribution (OoD) environments, motivated by the problem of cross-hospital generalization in critical care. We propose model-based and heuristic approaches to identify OoD environments and systematically compare models with different levels of held-out information. In particular, based on the assumption that models with access to OoD data should outperform other models, we train models across a range of experimental setups that include leave-one-hospital-out training and cross-sectional feature splits. We find that access to OoD data does not translate to increased performance, pointing to inherent limitations in defining potential OoD environments in the eICU Database potentially due to data harmonisation and sampling. Echoing similar results with other popular clinical benchmarks in the literature, new approaches are required to evaluate robust models in critical care.
Longitudinal cardio-respiratory fitness prediction through free-living wearable sensors
May 06, 2022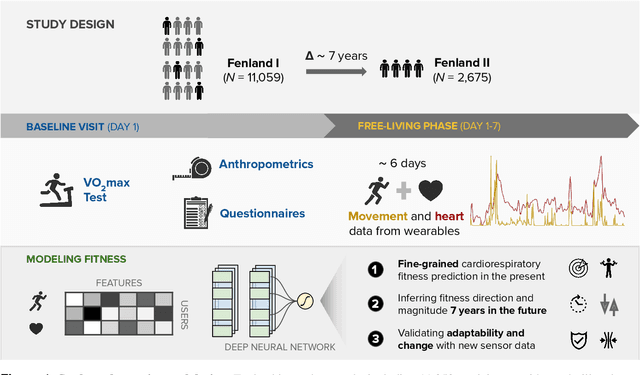
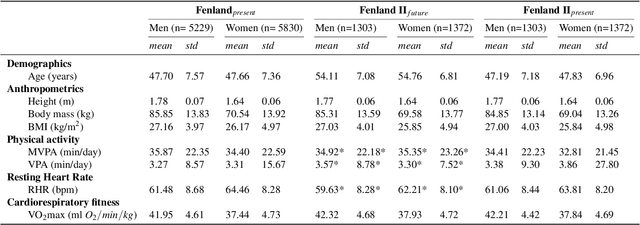
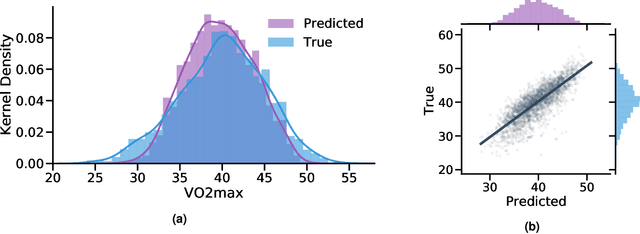
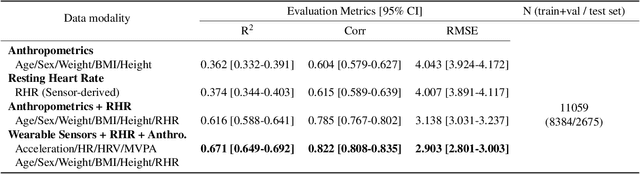
Abstract:Cardiorespiratory fitness is an established predictor of metabolic disease and mortality. Fitness is directly measured as maximal oxygen consumption (VO2max), or indirectly assessed using heart rate response to a standard exercise test. However, such testing is costly and burdensome, limiting its utility and scalability. Fitness can also be approximated using resting heart rate and self-reported exercise habits but with lower accuracy. Modern wearables capture dynamic heart rate data which, in combination with machine learning models, could improve fitness prediction. In this work, we analyze movement and heart rate signals from wearable sensors in free-living conditions from 11,059 participants who also underwent a standard exercise test, along with a longitudinal repeat cohort of 2,675 participants. We design algorithms and models that convert raw sensor data into cardio-respiratory fitness estimates, and validate these estimates' ability to capture fitness profiles in a longitudinal cohort over time while subjects engaged in real-world (non-exercise) behaviour. Additionally, we validate our methods with a third external cohort of 181 participants who underwent maximal VO2max testing, which is considered the gold standard measurement because it requires reaching one's maximum heart rate and exhaustion level. Our results show that the developed models yield a high correlation (r = 0.82, 95CI 0.80-0.83), when compared to the ground truth in a holdout sample. These models outperform conventional non-exercise fitness models and traditional bio-markers using measurements of normal daily living without the need for a specific exercise test. Additionally, we show the adaptability and applicability of this approach for detecting fitness change over time in the longitudinal subsample that repeated measurements after 7 years.
A Summary of the ComParE COVID-19 Challenges
Feb 17, 2022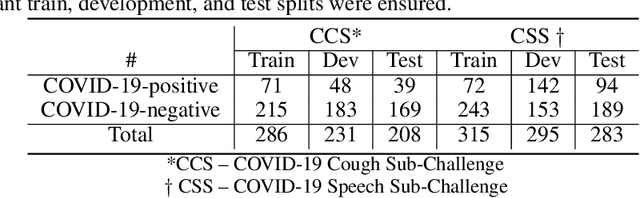
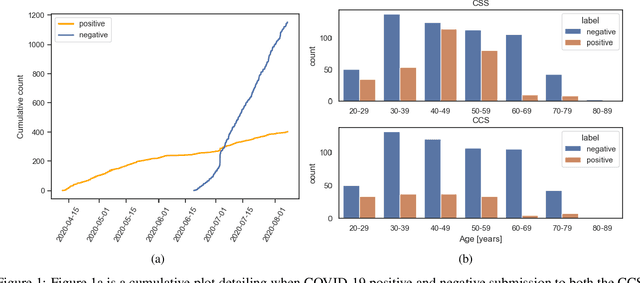
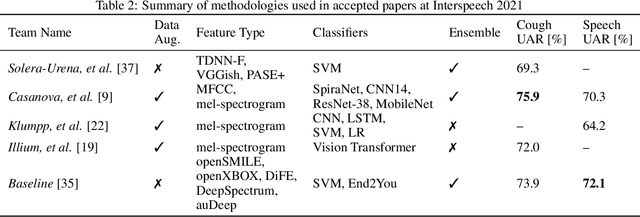
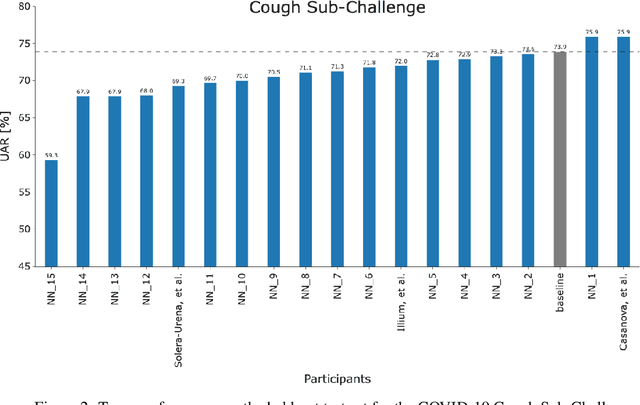
Abstract:The COVID-19 pandemic has caused massive humanitarian and economic damage. Teams of scientists from a broad range of disciplines have searched for methods to help governments and communities combat the disease. One avenue from the machine learning field which has been explored is the prospect of a digital mass test which can detect COVID-19 from infected individuals' respiratory sounds. We present a summary of the results from the INTERSPEECH 2021 Computational Paralinguistics Challenges: COVID-19 Cough, (CCS) and COVID-19 Speech, (CSS).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge