Dheeraj Mundhra
Variational Inference with Latent Space Quantization for Adversarial Resilience
Mar 24, 2019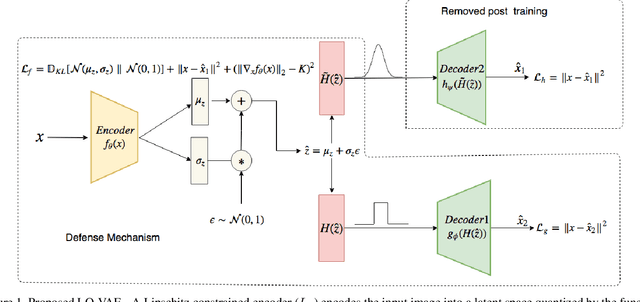
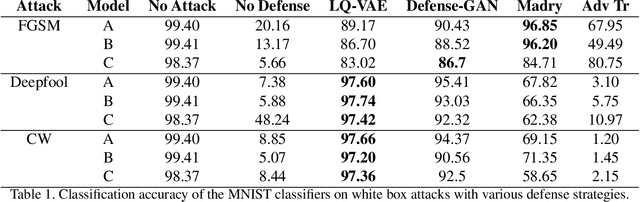
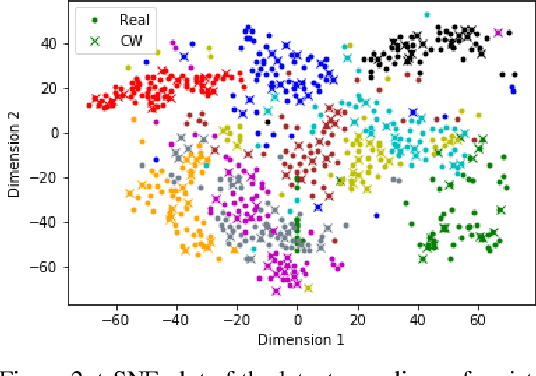
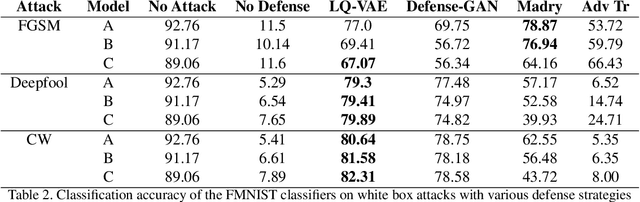
Abstract:Despite their tremendous success in modelling high-dimensional data manifolds, deep neural networks suffer from the threat of adversarial attacks - Existence of perceptually valid input-like samples obtained through careful perturbations that leads to degradation in the performance of underlying model. Major concerns with existing defense mechanisms include non-generalizability across different attacks, models and large inference time. In this paper, we propose a generalized defense mechanism capitalizing on the expressive power of regularized latent space based generative models. We design an adversarial filter, devoid of access to classifier and adversaries, which makes it usable in tandem with any classifier. The basic idea is to learn a Lipschitz constrained mapping from the data manifold, incorporating adversarial perturbations, to a quantized latent space and re-map it to the true data manifold. Specifically, we simultaneously auto-encode the data manifold and its perturbations implicitly through the perturbations of the regularized and quantized generative latent space, realized using variational inference. We demonstrate the efficacy of the proposed formulation in providing the resilience against multiple attack types (Black and white box) and methods, while being almost real-time. Our experiments show that the proposed method surpasses the state-of-the-art techniques in several cases.
Learning a Deep Convolution Network with Turing Test Adversaries for Microscopy Image Super Resolution
Jan 18, 2019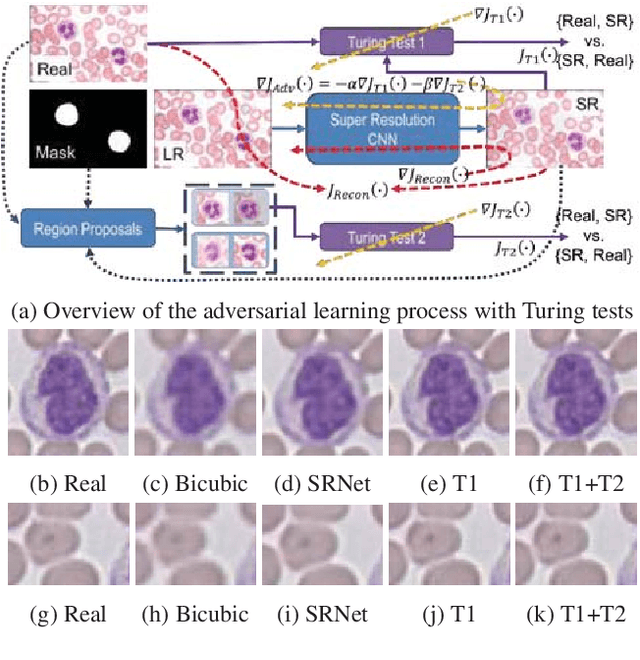
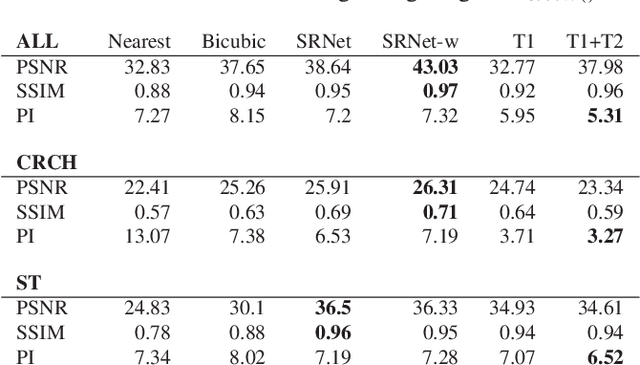
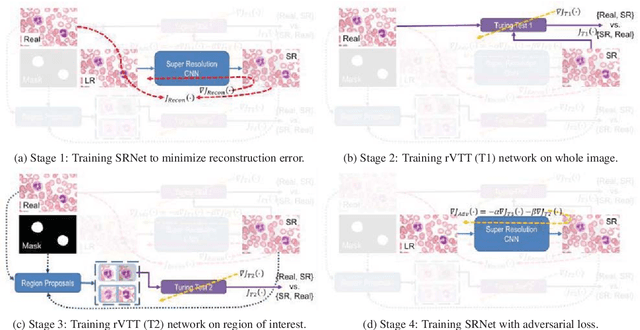

Abstract:Adversarially trained deep neural networks have significantly improved performance of single image super resolution, by hallucinating photorealistic local textures, thereby greatly reducing the perception difference between a real high resolution image and its super resolved (SR) counterpart. However, application to medical imaging requires preservation of diagnostically relevant features while refraining from introducing any diagnostically confusing artifacts. We propose using a deep convolutional super resolution network (SRNet) trained for (i) minimising reconstruction loss between the real and SR images, and (ii) maximally confusing learned relativistic visual Turing test (rVTT) networks to discriminate between (a) pair of real and SR images (T1) and (b) pair of patches in real and SR selected from region of interest (T2). The adversarial loss of T1 and T2 while backpropagated through SRNet helps it learn to reconstruct pathorealism in the regions of interest such as white blood cells (WBC) in peripheral blood smears or epithelial cells in histopathology of cancerous biopsy tissues, which are experimentally demonstrated here. Experiments performed for measuring signal distortion loss using peak signal to noise ratio (pSNR) and structural similarity (SSIM) with variation of SR scale factors, impact of rVTT adversarial losses, and impact on reporting using SR on a commercially available artificial intelligence (AI) digital pathology system substantiate our claims.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge