Dhanesh Ramachandram
From Features to Actions: Explainability in Traditional and Agentic AI Systems
Feb 09, 2026Abstract:Over the last decade, explainable AI has primarily focused on interpreting individual model predictions, producing post-hoc explanations that relate inputs to outputs under a fixed decision structure. Recent advances in large language models (LLMs) have enabled agentic AI systems whose behaviour unfolds over multi-step trajectories. In these settings, success and failure are determined by sequences of decisions rather than a single output. While useful, it remains unclear how explanation approaches designed for static predictions translate to agentic settings where behaviour emerges over time. In this work, we bridge the gap between static and agentic explainability by comparing attribution-based explanations with trace-based diagnostics across both settings. To make this distinction explicit, we empirically compare attribution-based explanations used in static classification tasks with trace-based diagnostics used in agentic benchmarks (TAU-bench Airline and AssistantBench). Our results show that while attribution methods achieve stable feature rankings in static settings (Spearman $ρ= 0.86$), they cannot be applied reliably to diagnose execution-level failures in agentic trajectories. In contrast, trace-grounded rubric evaluation for agentic settings consistently localizes behaviour breakdowns and reveals that state tracking inconsistency is 2.7$\times$ more prevalent in failed runs and reduces success probability by 49\%. These findings motivate a shift towards trajectory-level explainability for agentic systems when evaluating and diagnosing autonomous AI behaviour. Resources: https://github.com/VectorInstitute/unified-xai-evaluation-framework https://vectorinstitute.github.io/unified-xai-evaluation-framework
Interpretable Fine-Gray Deep Survival Model for Competing Risks: Predicting Post-Discharge Foot Complications for Diabetic Patients in Ontario
Nov 16, 2025Abstract:Model interpretability is crucial for establishing AI safety and clinician trust in medical applications for example, in survival modelling with competing risks. Recent deep learning models have attained very good predictive performance but their limited transparency, being black-box models, hinders their integration into clinical practice. To address this gap, we propose an intrinsically interpretable survival model called CRISPNAM-FG. Leveraging the structure of Neural Additive Models (NAMs) with separate projection vectors for each risk, our approach predicts the Cumulative Incidence Function using the Fine-Gray formulation, achieving high predictive power with intrinsically transparent and auditable predictions. We validated the model on several benchmark datasets and applied our model to predict future foot complications in diabetic patients across 29 Ontario hospitals (2016-2023). Our method achieves competitive performance compared to other deep survival models while providing transparency through shape functions and feature importance plots.
CRISP-NAM: Competing Risks Interpretable Survival Prediction with Neural Additive Models
May 27, 2025Abstract:Competing risks are crucial considerations in survival modelling, particularly in healthcare domains where patients may experience multiple distinct event types. We propose CRISP-NAM (Competing Risks Interpretable Survival Prediction with Neural Additive Models), an interpretable neural additive model for competing risks survival analysis which extends the neural additive architecture to model cause-specific hazards while preserving feature-level interpretability. Each feature contributes independently to risk estimation through dedicated neural networks, allowing for visualization of complex non-linear relationships between covariates and each competing risk. We demonstrate competitive performance on multiple datasets compared to existing approaches.
Bridging Electronic Health Records and Clinical Texts: Contrastive Learning for Enhanced Clinical Tasks
May 23, 2025Abstract:Conventional machine learning models, particularly tree-based approaches, have demonstrated promising performance across various clinical prediction tasks using electronic health record (EHR) data. Despite their strengths, these models struggle with tasks that require deeper contextual understanding, such as predicting 30-day hospital readmission. This can be primarily due to the limited semantic information available in structured EHR data. To address this limitation, we propose a deep multimodal contrastive learning (CL) framework that aligns the latent representations of structured EHR data with unstructured discharge summary notes. It works by pulling together paired EHR and text embeddings while pushing apart unpaired ones. Fine-tuning the pretrained EHR encoder extracted from this framework significantly boosts downstream task performance, e.g., a 4.1% AUROC enhancement over XGBoost for 30-day readmission prediction. Such results demonstrate the effect of integrating domain knowledge from clinical notes into EHR-based pipelines, enabling more accurate and context-aware clinical decision support systems.
Structure Optimization for Deep Multimodal Fusion Networks using Graph-Induced Kernels
Jul 03, 2017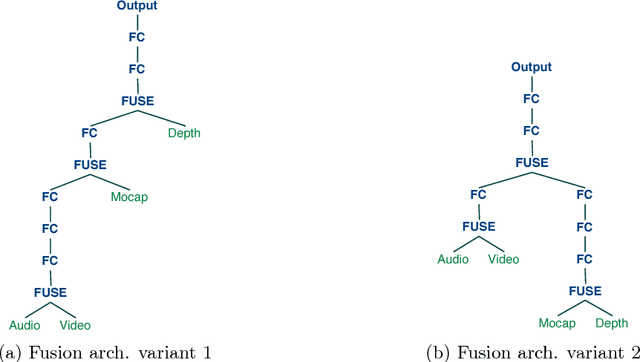
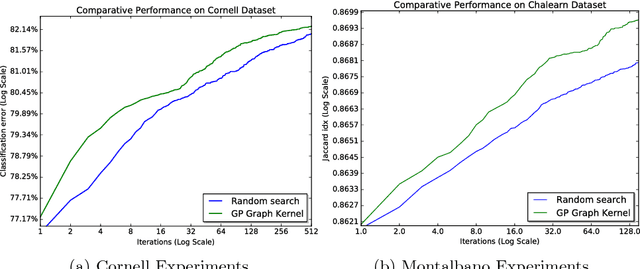
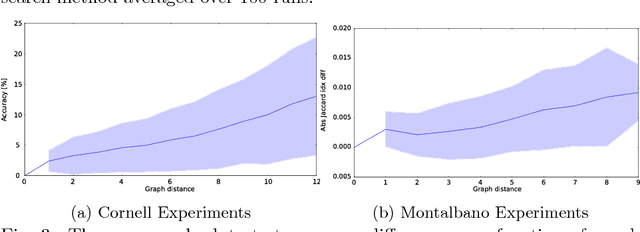
Abstract:A popular testbed for deep learning has been multimodal recognition of human activity or gesture involving diverse inputs such as video, audio, skeletal pose and depth images. Deep learning architectures have excelled on such problems due to their ability to combine modality representations at different levels of nonlinear feature extraction. However, designing an optimal architecture in which to fuse such learned representations has largely been a non-trivial human engineering effort. We treat fusion structure optimization as a hyper-parameter search and cast it as a discrete optimization problem under the Bayesian optimization framework. We propose a novel graph-induced kernel to compute structural similarities in the search space of tree-structured multimodal architectures and demonstrate its effectiveness using two challenging multimodal human activity recognition datasets.
LesionSeg: Semantic segmentation of skin lesions using Deep Convolutional Neural Network
Mar 15, 2017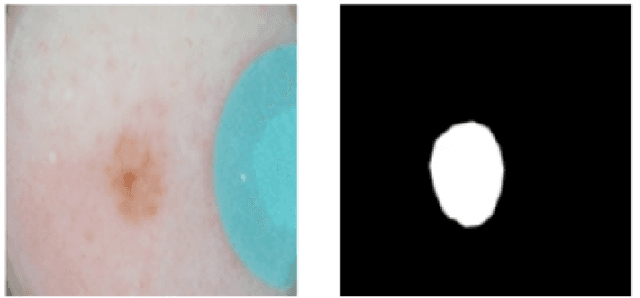
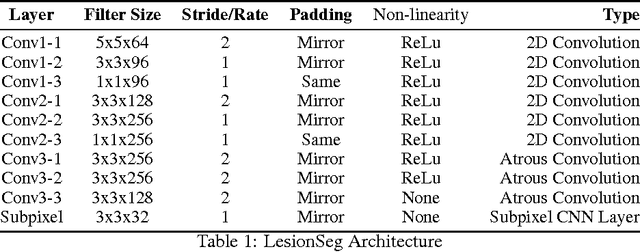

Abstract:We present a method for skin lesion segmentation for the ISIC 2017 Skin Lesion Segmentation Challenge. Our approach is based on a Fully Convolutional Network architecture which is trained end to end, from scratch, on a limited dataset. Our semantic segmentation architecture utilizes several recent innovations in particularly in the combined use of (i) use of atrous convolutions to increase the effective field of view of the network's receptive field without increasing the number of parameters, (ii) the use of network-in-network $1\times1$ convolution layers to add capacity to the network and (iii) state-of-art super-resolution upsampling of predictions using subpixel CNN layers. We reported a mean IOU score of 0.642 on the validation set provided by the organisers.
Skin Lesion Classification Using Deep Multi-scale Convolutional Neural Networks
Mar 04, 2017

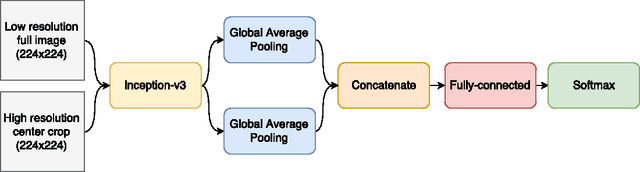
Abstract:We present a deep learning approach to the ISIC 2017 Skin Lesion Classification Challenge using a multi-scale convolutional neural network. Our approach utilizes an Inception-v3 network pre-trained on the ImageNet dataset, which is fine-tuned for skin lesion classification using two different scales of input images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge