David Rauber
Video Dataset for Surgical Phase, Keypoint, and Instrument Recognition in Laparoscopic Surgery (PhaKIR)
Nov 09, 2025Abstract:Robotic- and computer-assisted minimally invasive surgery (RAMIS) is increasingly relying on computer vision methods for reliable instrument recognition and surgical workflow understanding. Developing such systems often requires large, well-annotated datasets, but existing resources often address isolated tasks, neglect temporal dependencies, or lack multi-center variability. We present the Surgical Procedure Phase, Keypoint, and Instrument Recognition (PhaKIR) dataset, comprising eight complete laparoscopic cholecystectomy videos recorded at three medical centers. The dataset provides frame-level annotations for three interconnected tasks: surgical phase recognition (485,875 frames), instrument keypoint estimation (19,435 frames), and instrument instance segmentation (19,435 frames). PhaKIR is, to our knowledge, the first multi-institutional dataset to jointly provide phase labels, instrument pose information, and pixel-accurate instrument segmentations, while also enabling the exploitation of temporal context since full surgical procedure sequences are available. It served as the basis for the PhaKIR Challenge as part of the Endoscopic Vision (EndoVis) Challenge at MICCAI 2024 to benchmark methods in surgical scene understanding, thereby further validating the dataset's quality and relevance. The dataset is publicly available upon request via the Zenodo platform.
Comparative validation of surgical phase recognition, instrument keypoint estimation, and instrument instance segmentation in endoscopy: Results of the PhaKIR 2024 challenge
Jul 22, 2025Abstract:Reliable recognition and localization of surgical instruments in endoscopic video recordings are foundational for a wide range of applications in computer- and robot-assisted minimally invasive surgery (RAMIS), including surgical training, skill assessment, and autonomous assistance. However, robust performance under real-world conditions remains a significant challenge. Incorporating surgical context - such as the current procedural phase - has emerged as a promising strategy to improve robustness and interpretability. To address these challenges, we organized the Surgical Procedure Phase, Keypoint, and Instrument Recognition (PhaKIR) sub-challenge as part of the Endoscopic Vision (EndoVis) challenge at MICCAI 2024. We introduced a novel, multi-center dataset comprising thirteen full-length laparoscopic cholecystectomy videos collected from three distinct medical institutions, with unified annotations for three interrelated tasks: surgical phase recognition, instrument keypoint estimation, and instrument instance segmentation. Unlike existing datasets, ours enables joint investigation of instrument localization and procedural context within the same data while supporting the integration of temporal information across entire procedures. We report results and findings in accordance with the BIAS guidelines for biomedical image analysis challenges. The PhaKIR sub-challenge advances the field by providing a unique benchmark for developing temporally aware, context-driven methods in RAMIS and offers a high-quality resource to support future research in surgical scene understanding.
OpenMIBOOD: Open Medical Imaging Benchmarks for Out-Of-Distribution Detection
Mar 20, 2025



Abstract:The growing reliance on Artificial Intelligence (AI) in critical domains such as healthcare demands robust mechanisms to ensure the trustworthiness of these systems, especially when faced with unexpected or anomalous inputs. This paper introduces the Open Medical Imaging Benchmarks for Out-Of-Distribution Detection (OpenMIBOOD), a comprehensive framework for evaluating out-of-distribution (OOD) detection methods specifically in medical imaging contexts. OpenMIBOOD includes three benchmarks from diverse medical domains, encompassing 14 datasets divided into covariate-shifted in-distribution, near-OOD, and far-OOD categories. We evaluate 24 post-hoc methods across these benchmarks, providing a standardized reference to advance the development and fair comparison of OOD detection methods. Results reveal that findings from broad-scale OOD benchmarks in natural image domains do not translate to medical applications, underscoring the critical need for such benchmarks in the medical field. By mitigating the risk of exposing AI models to inputs outside their training distribution, OpenMIBOOD aims to support the advancement of reliable and trustworthy AI systems in healthcare. The repository is available at https://github.com/remic-othr/OpenMIBOOD.
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020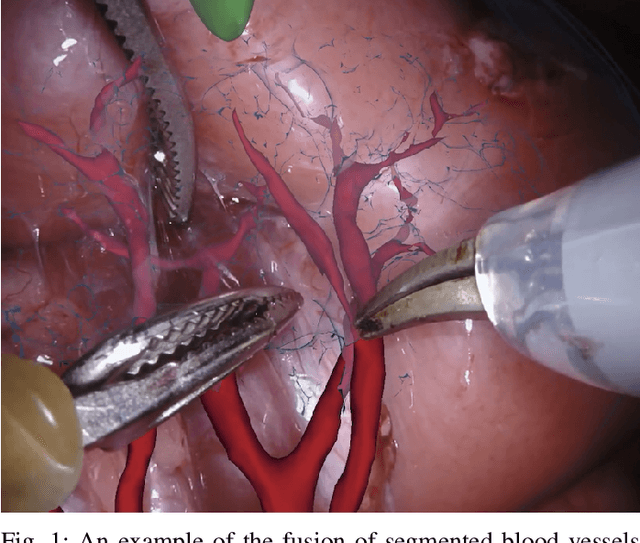
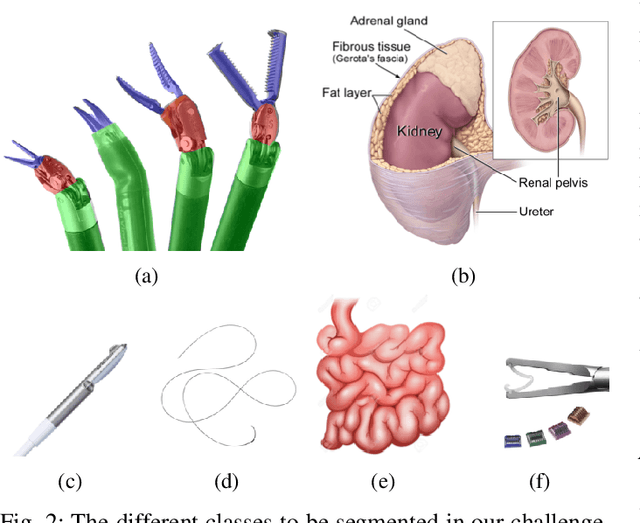
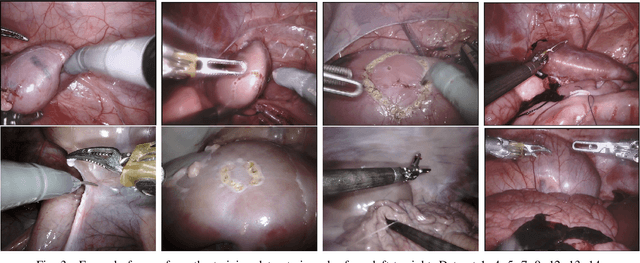
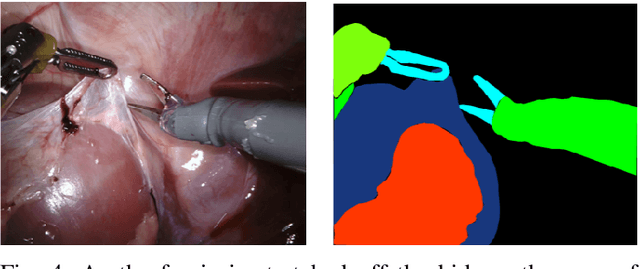
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge