Daniel Rubin
Biologic and Prognostic Feature Scores from Whole-Slide Histology Images Using Deep Learning
Oct 24, 2019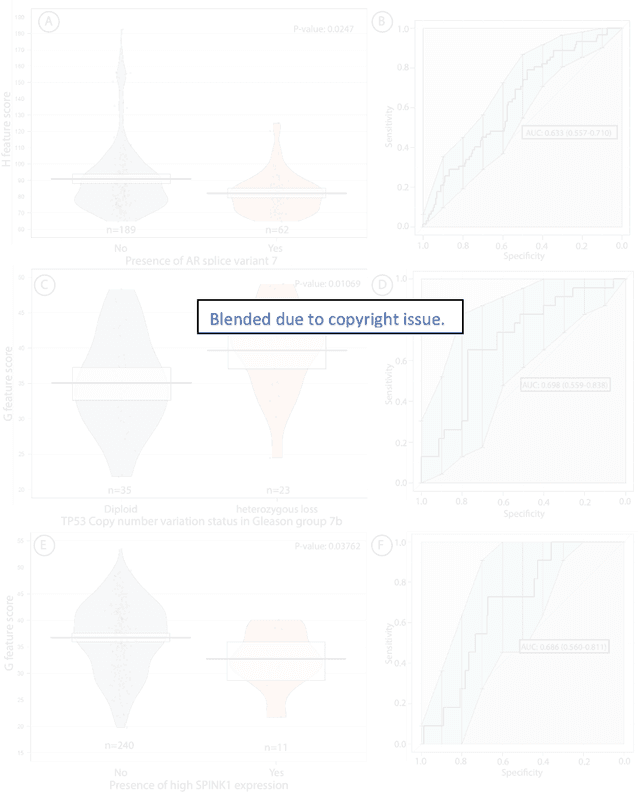
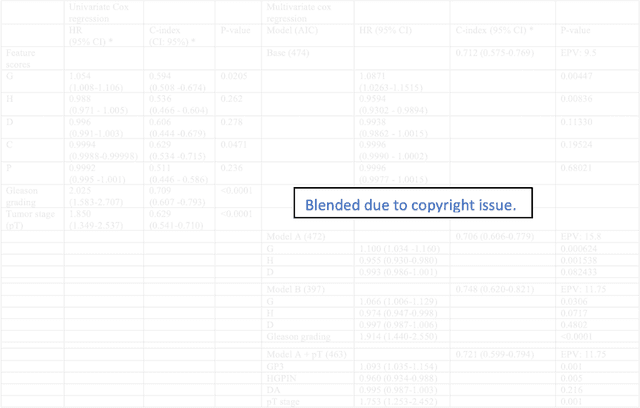
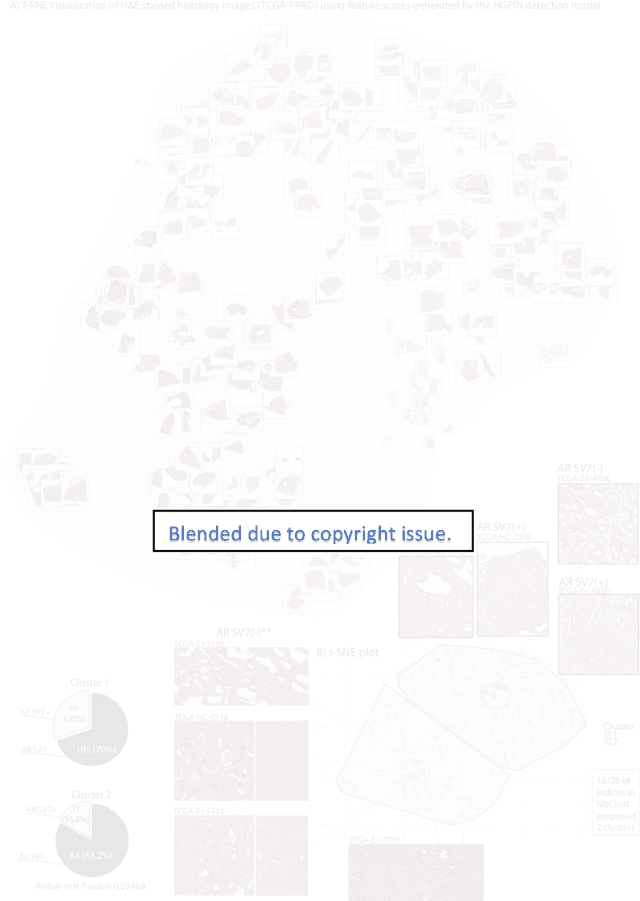
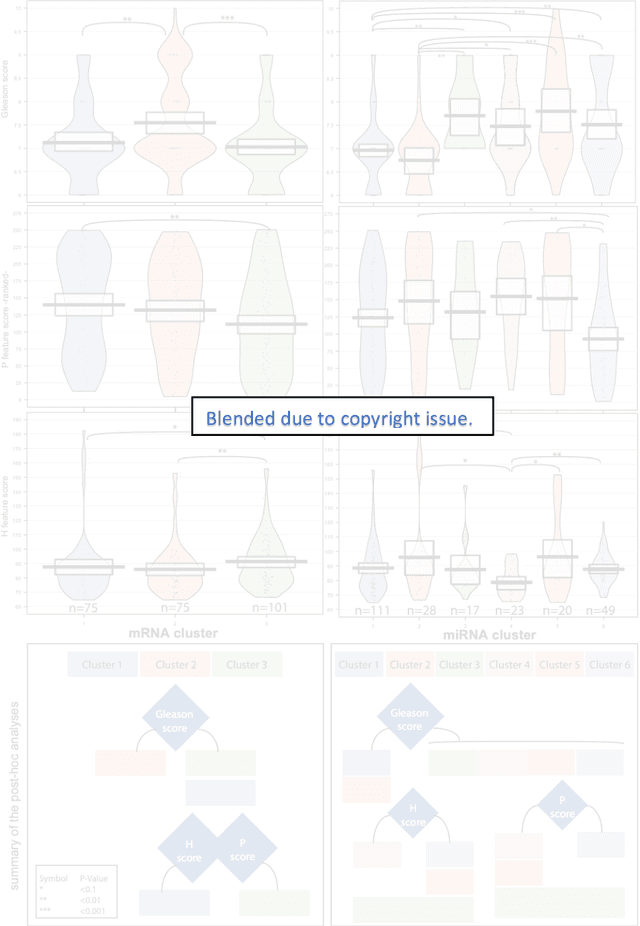
Abstract:Histopathology is a reflection of the molecular changes and provides prognostic phenotypes representing the disease progression. In this study, we introduced feature scores generated from hematoxylin and eosin histology images based on deep learning (DL) models developed for prostate pathology. We demonstrated that these feature scores were significantly prognostic for time to event endpoints (biochemical recurrence and cancer-specific survival) and had simultaneously molecular biologic associations to relevant genomic alterations and molecular subtypes using already trained DL models that were not previously exposed to the datasets of the current study. Further, we discussed the potential of such feature scores to improve the current tumor grading system and the challenges that are associated with tumor heterogeneity and the development of prognostic models from histology images. Our findings uncover the potential of feature scores from histology images as digital biomarkers in precision medicine and as an expanding utility for digital pathology.
Deep Learning for Prostate Pathology
Oct 16, 2019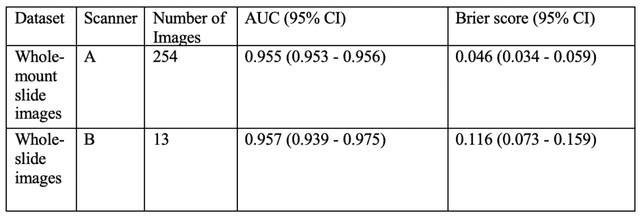
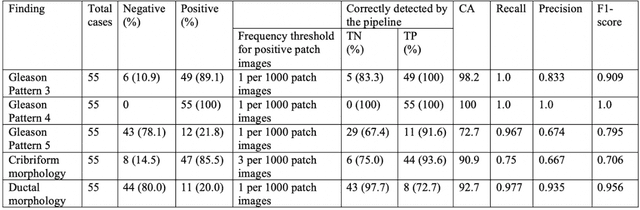
Abstract:The current study detects different morphologies related to prostate pathology using deep learning models; these models were evaluated on 2,121 hematoxylin and eosin (H&E) stain histology images captured using bright field microscopy, which spanned a variety of image qualities, origins (whole slide, tissue micro array, whole mount, Internet), scanning machines, timestamps, H&E staining protocols, and institutions. For case usage, these models were applied for the annotation tasks in clinician-oriented pathology reports for prostatectomy specimens. The true positive rate (TPR) for slides with prostate cancer was 99.7% by a false positive rate of 0.785%. The F1-scores of Gleason patterns reported in pathology reports ranged from 0.795 to 1.0 at the case level. TPR was 93.6% for the cribriform morphology and 72.6% for the ductal morphology. The correlation between the ground truth and the prediction for the relative tumor volume was 0.987 n. Our models cover the major components of prostate pathology and successfully accomplish the annotation tasks.
Deep Active Lesion Segmentation
Aug 21, 2019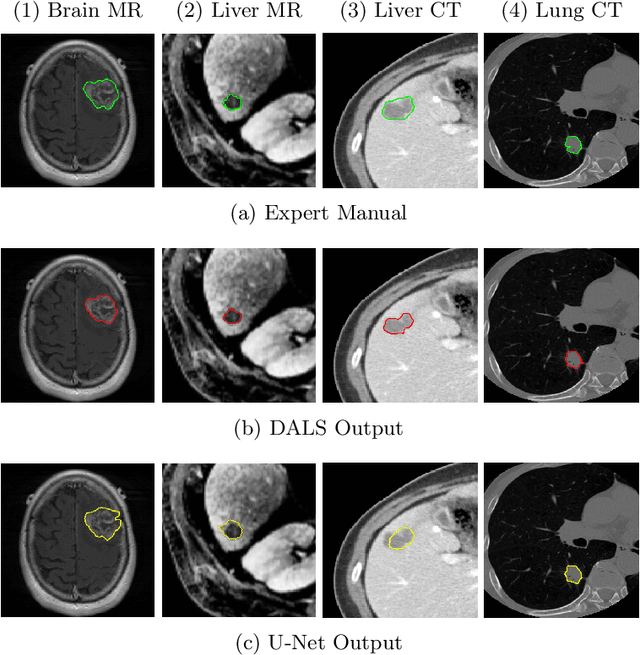
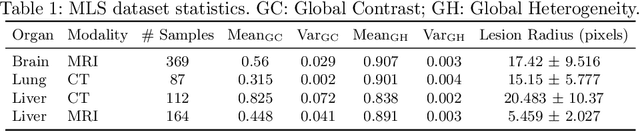
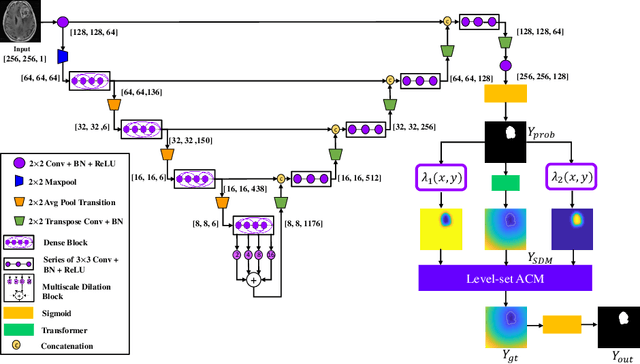
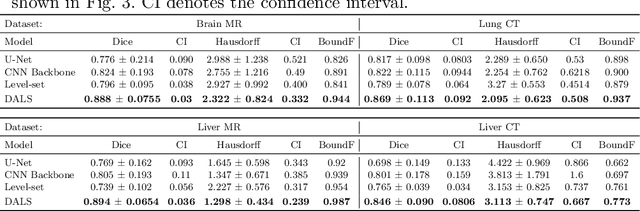
Abstract:Lesion segmentation is an important problem in computer-assisted diagnosis that remains challenging due to the prevalence of low contrast, irregular boundaries that are unamenable to shape priors. We introduce Deep Active Lesion Segmentation (DALS), a fully automated segmentation framework for that leverages the powerful nonlinear feature extraction abilities of fully Convolutional Neural Networks (CNNs) and the precise boundary delineation abilities of Active Contour Models (ACMs). Our DALS framework benefits from an improved level-set ACM formulation with a per-pixel-parameterized energy functional and a novel multiscale encoder-decoder CNN that learns an initialization probability map along with parameter maps for the ACM. We evaluate our lesion segmentation model on a new Multiorgan Lesion Segmentation (MLS) dataset that contains images of various organs, including brain, liver, and lung, across different imaging modalities---MR and CT. Our results demonstrate favorable performance compared to competing methods, especially for small training datasets.
* Accepted to Machine Learning in Medical Imaging (MLMI 2019)
Cross-Modal Data Programming Enables Rapid Medical Machine Learning
Mar 26, 2019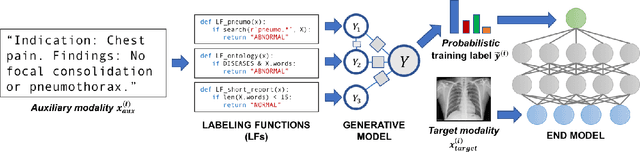
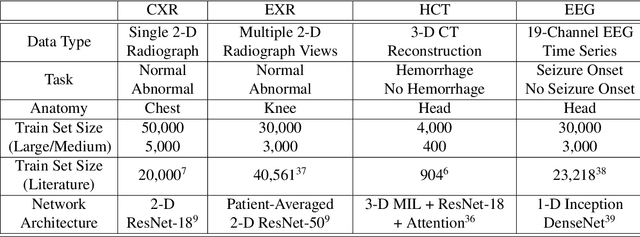
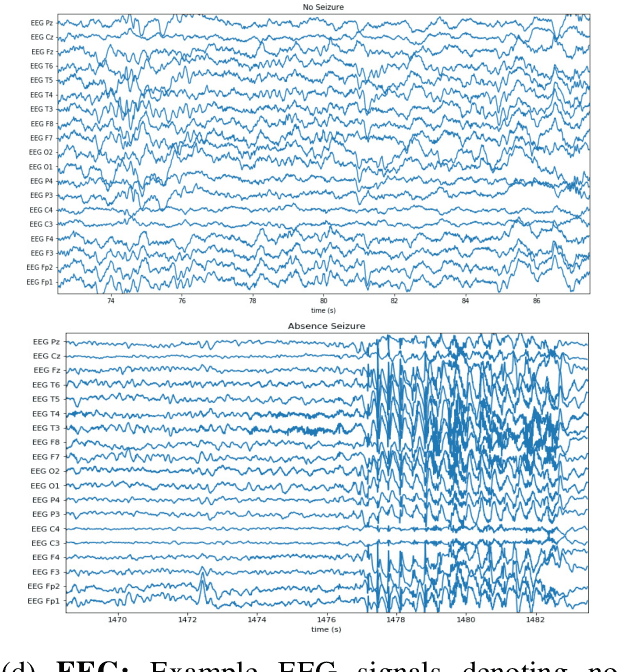
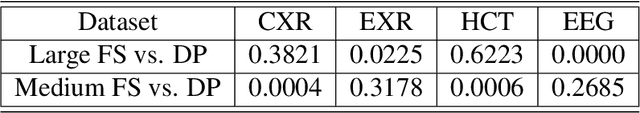
Abstract:Labeling training datasets has become a key barrier to building medical machine learning models. One strategy is to generate training labels programmatically, for example by applying natural language processing pipelines to text reports associated with imaging studies. We propose cross-modal data programming, which generalizes this intuitive strategy in a theoretically-grounded way that enables simpler, clinician-driven input, reduces required labeling time, and improves with additional unlabeled data. In this approach, clinicians generate training labels for models defined over a target modality (e.g. images or time series) by writing rules over an auxiliary modality (e.g. text reports). The resulting technical challenge consists of estimating the accuracies and correlations of these rules; we extend a recent unsupervised generative modeling technique to handle this cross-modal setting in a provably consistent way. Across four applications in radiography, computed tomography, and electroencephalography, and using only several hours of clinician time, our approach matches or exceeds the efficacy of physician-months of hand-labeling with statistical significance, demonstrating a fundamentally faster and more flexible way of building machine learning models in medicine.
A Deep-learning Approach for Prognosis of Age-Related Macular Degeneration Disease using SD-OCT Imaging Biomarkers
Feb 27, 2019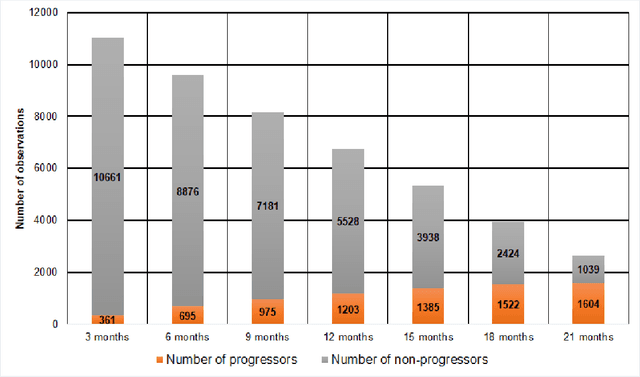
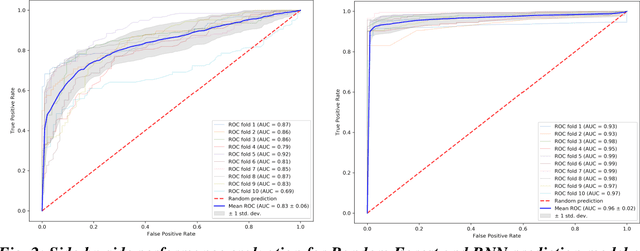
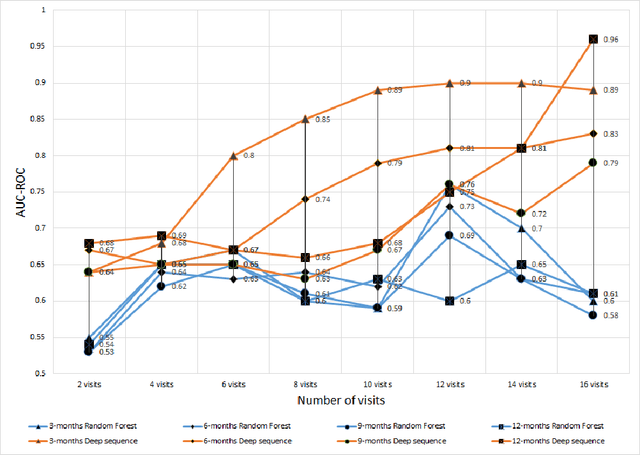
Abstract:We propose a hybrid sequential deep learning model to predict the risk of AMD progression in non-exudative AMD eyes at multiple timepoints, starting from short-term progression (3-months) up to long-term progression (21-months). Proposed model combines radiomics and deep learning to handle challenges related to imperfect ratio of OCT scan dimension and training cohort size. We considered a retrospective clinical trial dataset that includes 671 fellow eyes with 13,954 dry AMD observations for training and validating the machine learning models on a 10-fold cross validation setting. The proposed RNN model achieved high accuracy (0.96 AUCROC) for the prediction of both short term and long-term AMD progression, and outperformed the traditional random forest model trained. High accuracy achieved by the RNN establishes the ability to identify AMD patients at risk of progressing to advanced AMD at an early stage which could have a high clinical impact as it allows for optimal clinical follow-up, with more frequent screening and potential earlier treatment for those patients at high risk.
TVAE: Triplet-Based Variational Autoencoder using Metric Learning
Apr 03, 2018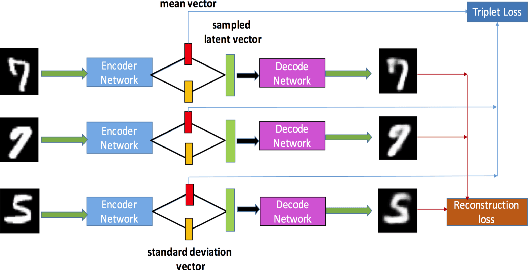
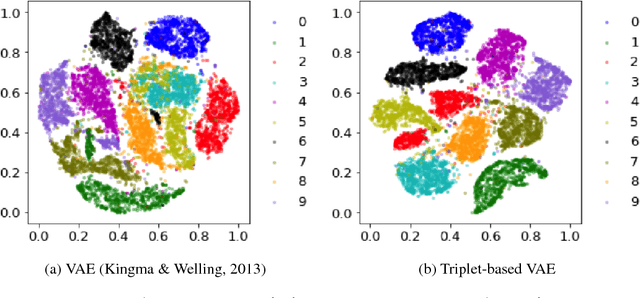

Abstract:Deep metric learning has been demonstrated to be highly effective in learning semantic representation and encoding information that can be used to measure data similarity, by relying on the embedding learned from metric learning. At the same time, variational autoencoder (VAE) has widely been used to approximate inference and proved to have a good performance for directed probabilistic models. However, for traditional VAE, the data label or feature information are intractable. Similarly, traditional representation learning approaches fail to represent many salient aspects of the data. In this project, we propose a novel integrated framework to learn latent embedding in VAE by incorporating deep metric learning. The features are learned by optimizing a triplet loss on the mean vectors of VAE in conjunction with standard evidence lower bound (ELBO) of VAE. This approach, which we call Triplet based Variational Autoencoder (TVAE), allows us to capture more fine-grained information in the latent embedding. Our model is tested on MNIST data set and achieves a high triplet accuracy of 95.60% while the traditional VAE (Kingma & Welling, 2013) achieves triplet accuracy of 75.08%.
Optimizing and Visualizing Deep Learning for Benign/Malignant Classification in Breast Tumors
May 17, 2017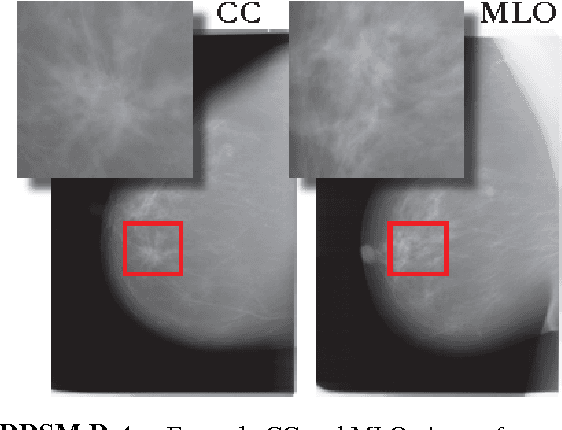
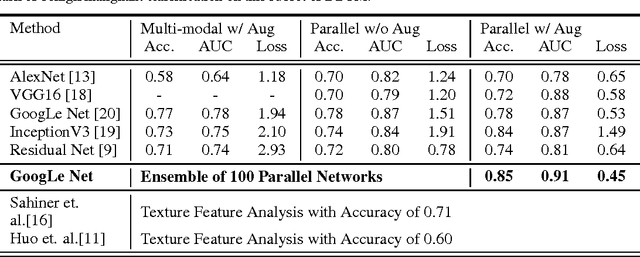
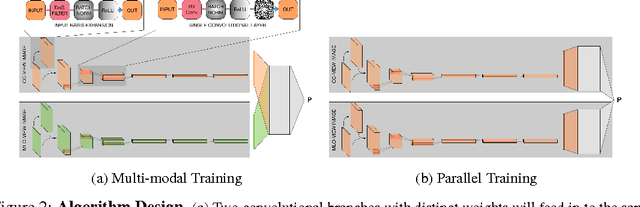
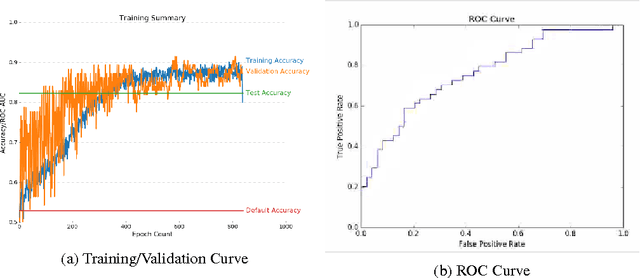
Abstract:Breast cancer has the highest incidence and second highest mortality rate for women in the US. Our study aims to utilize deep learning for benign/malignant classification of mammogram tumors using a subset of cases from the Digital Database of Screening Mammography (DDSM). Though it was a small dataset from the view of Deep Learning (about 1000 patients), we show that currently state of the art architectures of deep learning can find a robust signal, even when trained from scratch. Using convolutional neural networks (CNNs), we are able to achieve an accuracy of 85% and an ROC AUC of 0.91, while leading hand-crafted feature based methods are only able to achieve an accuracy of 71%. We investigate an amalgamation of architectures to show that our best result is reached with an ensemble of the lightweight GoogLe Nets tasked with interpreting both the coronal caudal view and the mediolateral oblique view, simply averaging the probability scores of both views to make the final prediction. In addition, we have created a novel method to visualize what features the neural network detects for the benign/malignant classification, and have correlated those features with well known radiological features, such as spiculation. Our algorithm significantly improves existing classification methods for mammography lesions and identifies features that correlate with established clinical markers.
Classification of Hepatic Lesions using the Matching Metric
Oct 02, 2012
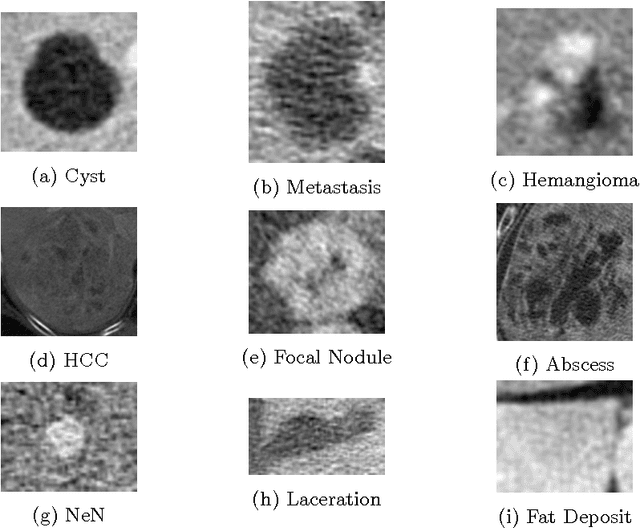

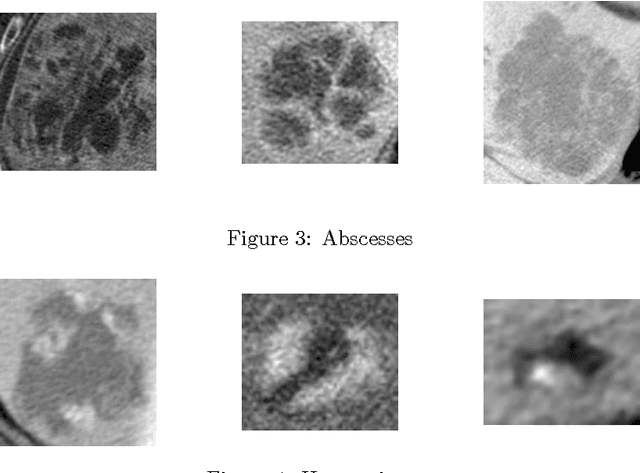
Abstract:In this paper we present a methodology of classifying hepatic (liver) lesions using multidimensional persistent homology, the matching metric (also called the bottleneck distance), and a support vector machine. We present our classification results on a dataset of 132 lesions that have been outlined and annotated by radiologists. We find that topological features are useful in the classification of hepatic lesions. We also find that two-dimensional persistent homology outperforms one-dimensional persistent homology in this application.
A Hybrid Method for Distance Metric Learning
Jun 29, 2012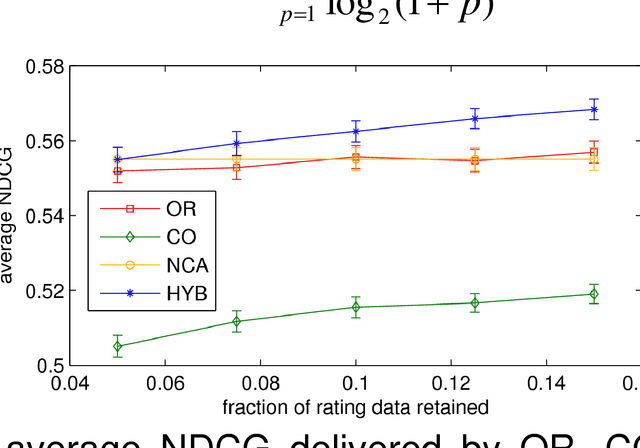
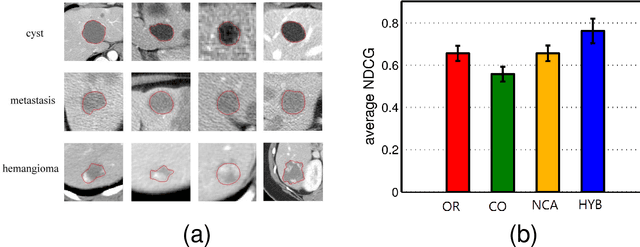
Abstract:We consider the problem of learning a measure of distance among vectors in a feature space and propose a hybrid method that simultaneously learns from similarity ratings assigned to pairs of vectors and class labels assigned to individual vectors. Our method is based on a generative model in which class labels can provide information that is not encoded in feature vectors but yet relates to perceived similarity between objects. Experiments with synthetic data as well as a real medical image retrieval problem demonstrate that leveraging class labels through use of our method improves retrieval performance significantly.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge