Benjamin Nye
Towards AI-Assisted Generation of Military Training Scenarios
Nov 10, 2025Abstract:Achieving expert-level performance in simulation-based training relies on the creation of complex, adaptable scenarios, a traditionally laborious and resource intensive process. Although prior research explored scenario generation for military training, pre-LLM AI tools struggled to generate sufficiently complex or adaptable scenarios. This paper introduces a multi-agent, multi-modal reasoning framework that leverages Large Language Models (LLMs) to generate critical training artifacts, such as Operations Orders (OPORDs). We structure our framework by decomposing scenario generation into a hierarchy of subproblems, and for each one, defining the role of the AI tool: (1) generating options for a human author to select from, (2) producing a candidate product for human approval or modification, or (3) generating textual artifacts fully automatically. Our framework employs specialized LLM-based agents to address distinct subproblems. Each agent receives input from preceding subproblem agents, integrating both text-based scenario details and visual information (e.g., map features, unit positions and applies specialized reasoning to produce appropriate outputs. Subsequent agents process these outputs sequentially, preserving logical consistency and ensuring accurate document generation. This multi-agent strategy overcomes the limitations of basic prompting or single-agent approaches when tackling such highly complex tasks. We validate our framework through a proof-of-concept that generates the scheme of maneuver and movement section of an OPORD while estimating map positions and movements as a precursor demonstrating its feasibility and accuracy. Our results demonstrate the potential of LLM-driven multi-agent systems to generate coherent, nuanced documents and adapt dynamically to changing conditions, advancing automation in scenario generation for military training.
From Videos to Indexed Knowledge Graphs -- Framework to Marry Methods for Multimodal Content Analysis and Understanding
Oct 01, 2025Abstract:Analysis of multi-modal content can be tricky, computationally expensive, and require a significant amount of engineering efforts. Lots of work with pre-trained models on static data is out there, yet fusing these opensource models and methods with complex data such as videos is relatively challenging. In this paper, we present a framework that enables efficiently prototyping pipelines for multi-modal content analysis. We craft a candidate recipe for a pipeline, marrying a set of pre-trained models, to convert videos into a temporal semi-structured data format. We translate this structure further to a frame-level indexed knowledge graph representation that is query-able and supports continual learning, enabling the dynamic incorporation of new domain-specific knowledge through an interactive medium.
Fine-tuning for Better Few Shot Prompting: An Empirical Comparison for Short Answer Grading
Aug 06, 2025Abstract:Research to improve Automated Short Answer Grading has recently focused on Large Language Models (LLMs) with prompt engineering and no- or few-shot prompting to achieve best results. This is in contrast to the fine-tuning approach, which has historically required large-scale compute clusters inaccessible to most users. New closed-model approaches such as OpenAI's fine-tuning service promise results with as few as 100 examples, while methods using open weights such as quantized low-rank adaptive (QLORA) can be used to fine-tune models on consumer GPUs. We evaluate both of these fine-tuning methods, measuring their interaction with few-shot prompting for automated short answer grading (ASAG) with structured (JSON) outputs. Our results show that finetuning with small amounts of data has limited utility for Llama open-weight models, but that fine-tuning methods can outperform few-shot baseline instruction-tuned LLMs for OpenAI's closed models. While our evaluation set is limited, we find some evidence that the observed benefits of finetuning may be impacted by the domain subject matter. Lastly, we observed dramatic improvement with the LLama 3.1 8B-Instruct open-weight model by seeding the initial training examples with a significant amount of cheaply generated synthetic training data.
Jointly Extracting Interventions, Outcomes, and Findings from RCT Reports with LLMs
May 05, 2023



Abstract:Results from Randomized Controlled Trials (RCTs) establish the comparative effectiveness of interventions, and are in turn critical inputs for evidence-based care. However, results from RCTs are presented in (often unstructured) natural language articles describing the design, execution, and outcomes of trials; clinicians must manually extract findings pertaining to interventions and outcomes of interest from such articles. This onerous manual process has motivated work on (semi-)automating extraction of structured evidence from trial reports. In this work we propose and evaluate a text-to-text model built on instruction-tuned Large Language Models (LLMs) to jointly extract Interventions, Outcomes, and Comparators (ICO elements) from clinical abstracts, and infer the associated results reported. Manual (expert) and automated evaluations indicate that framing evidence extraction as a conditional generation task and fine-tuning LLMs for this purpose realizes considerable ($\sim$20 point absolute F1 score) gains over the previous SOTA. We perform ablations and error analyses to assess aspects that contribute to model performance, and to highlight potential directions for further improvements. We apply our model to a collection of published RCTs through mid-2022, and release a searchable database of structured findings (anonymously for now): bit.ly/joint-relations-extraction-mlhc
A Corpus with Multi-Level Annotations of Patients, Interventions and Outcomes to Support Language Processing for Medical Literature
Jun 11, 2018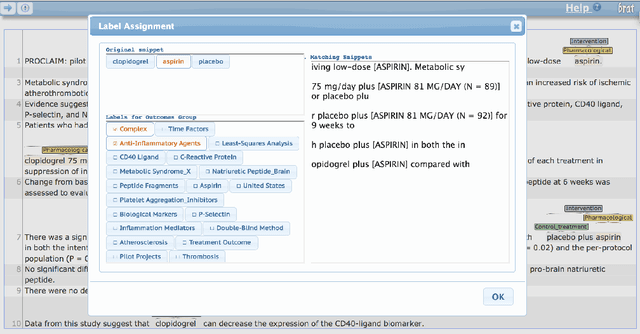

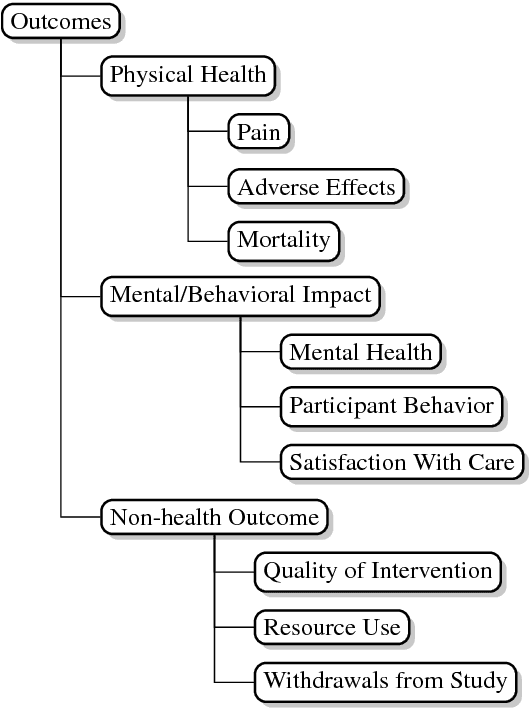
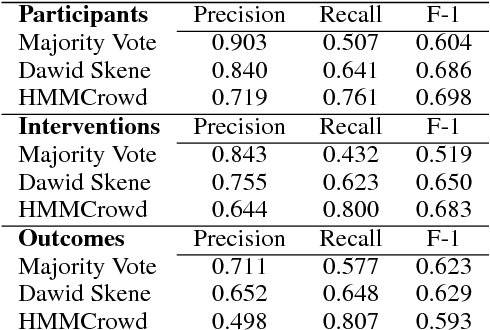
Abstract:We present a corpus of 5,000 richly annotated abstracts of medical articles describing clinical randomized controlled trials. Annotations include demarcations of text spans that describe the Patient population enrolled, the Interventions studied and to what they were Compared, and the Outcomes measured (the `PICO' elements). These spans are further annotated at a more granular level, e.g., individual interventions within them are marked and mapped onto a structured medical vocabulary. We acquired annotations from a diverse set of workers with varying levels of expertise and cost. We describe our data collection process and the corpus itself in detail. We then outline a set of challenging NLP tasks that would aid searching of the medical literature and the practice of evidence-based medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge