Get our free extension to see links to code for papers anywhere online!Free add-on: code for papers everywhere!Free add-on: See code for papers anywhere!
Ben Cocanougher
Semiparametric spectral modeling of the Drosophila connectome
May 09, 2017Authors:Carey E. Priebe, Youngser Park, Minh Tang, Avanti Athreya, Vince Lyzinski, Joshua T. Vogelstein, Yichen Qin, Ben Cocanougher, Katharina Eichler, Marta Zlatic(+1 more)
Figures and Tables: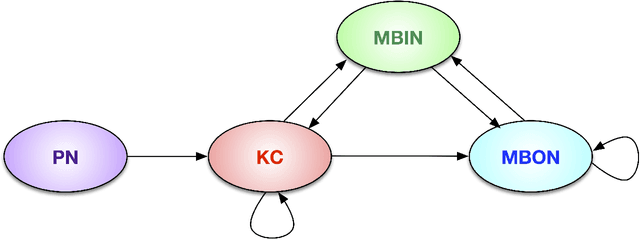
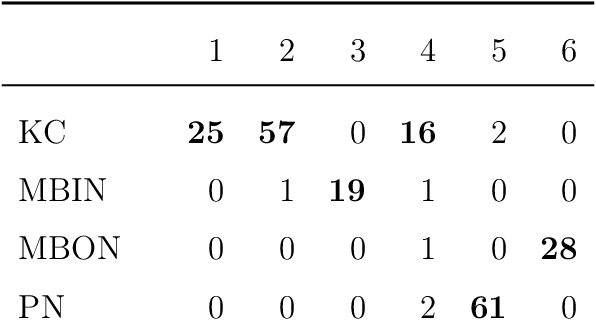
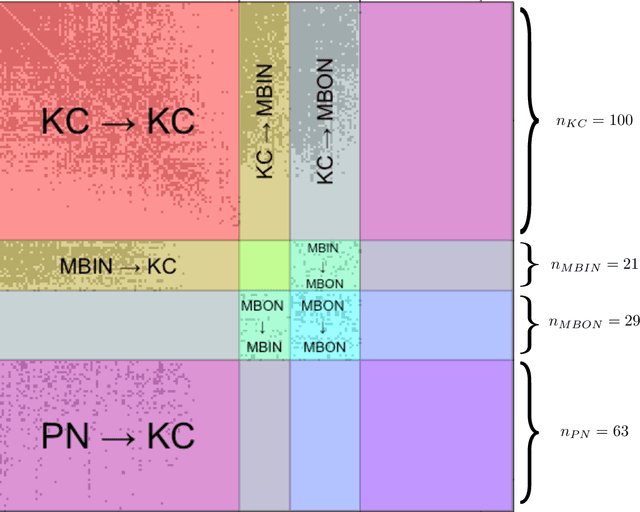
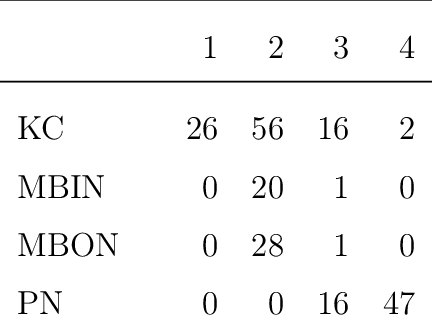




Abstract:We present semiparametric spectral modeling of the complete larval Drosophila mushroom body connectome. Motivated by a thorough exploratory data analysis of the network via Gaussian mixture modeling (GMM) in the adjacency spectral embedding (ASE) representation space, we introduce the latent structure model (LSM) for network modeling and inference. LSM is a generalization of the stochastic block model (SBM) and a special case of the random dot product graph (RDPG) latent position model, and is amenable to semiparametric GMM in the ASE representation space. The resulting connectome code derived via semiparametric GMM composed with ASE captures latent connectome structure and elucidates biologically relevant neuronal properties.
Via
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge