Babak Taati
Concurrent Validity of Automatic Speech and Pause Measures During Passage Reading in ALS
Aug 22, 2022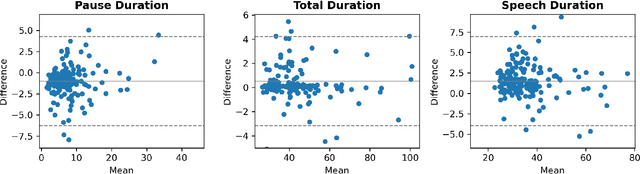
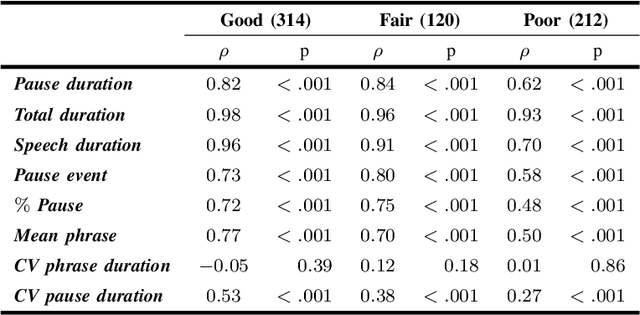
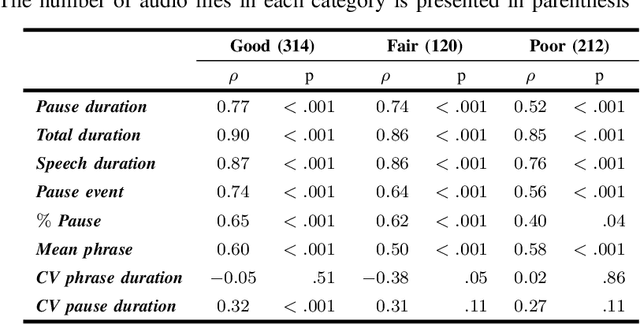
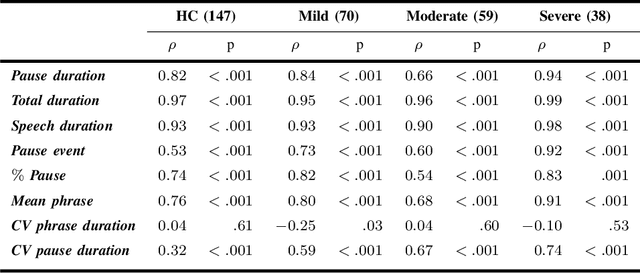
Abstract:The analysis of speech measures in individuals with amyotrophic lateral sclerosis (ALS) can provide essential information for early diagnosis and tracking disease progression. However, current methods for extracting speech and pause features are manual or semi-automatic, which makes them time-consuming and labour-intensive. The advent of speech-text alignment algorithms provides an opportunity for inexpensive, automated, and accurate analysis of speech measures in individuals with ALS. There is a need to validate speech and pause features calculated by these algorithms against current gold standard methods. In this study, we extracted 8 speech/pause features from 646 audio files of individuals with ALS and healthy controls performing passage reading. Two pretrained forced alignment models - one using transformers and another using a Gaussian mixture / hidden Markov architecture - were used for automatic feature extraction. The results were then validated against semi-automatic speech/pause analysis software, with further subgroup analyses based on audio quality and disease severity. Features extracted using transformer-based forced alignment had the highest agreement with gold standards, including in terms of audio quality and disease severity. This study lays the groundwork for future intelligent diagnostic support systems for clinicians, and for novel methods of tracking disease progression remotely from home.
Automated Temporal Segmentation of Orofacial Assessment Videos
Aug 22, 2022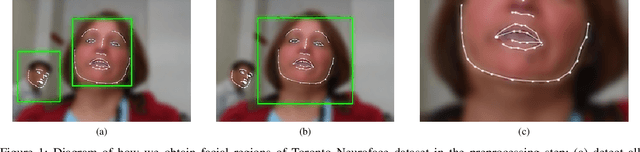
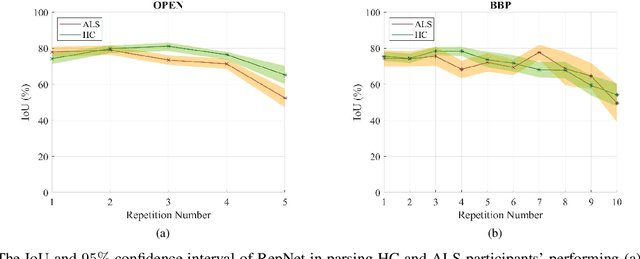
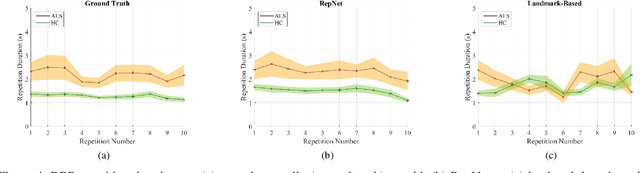
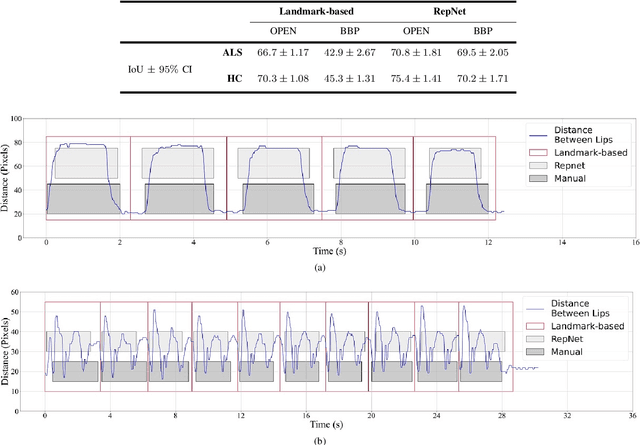
Abstract:Computer vision techniques can help automate or partially automate clinical examination of orofacial impairments to provide accurate and objective assessments. Towards the development of such automated systems, we evaluated two approaches to detect and temporally segment (parse) repetitions in orofacial assessment videos. Recorded videos of participants with amyotrophic lateral sclerosis (ALS) and healthy control (HC) individuals were obtained from the Toronto NeuroFace Dataset. Two approaches for repetition detection and parsing were examined: one based on engineered features from tracked facial landmarks and peak detection in the distance between the vermilion-cutaneous junction of the upper and lower lips (baseline analysis), and another using a pre-trained transformer-based deep learning model called RepNet (Dwibedi et al, 2020), which automatically detects periodicity, and parses periodic and semi-periodic repetitions in video data. In experimental evaluation of two orofacial assessments tasks, - repeating maximum mouth opening (OPEN) and repeating the sentence "Buy Bobby a Puppy" (BBP) - RepNet provided better parsing than the landmark-based approach, quantified by higher mean intersection-over-union (IoU) with respect to ground truth manual parsing. Automated parsing using RepNet also clearly separated HC and ALS participants based on the duration of BBP repetitions, whereas the landmark-based method could not.
Estimating Parkinsonism Severity in Natural Gait Videos of Older Adults with Dementia
May 07, 2021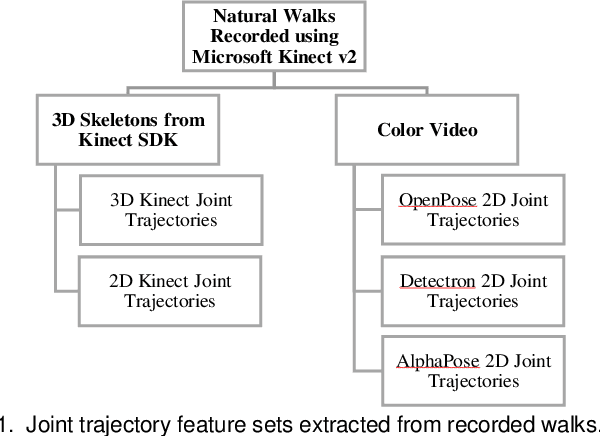

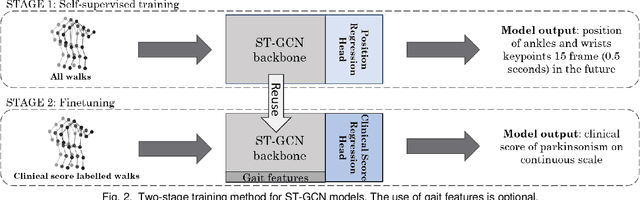
Abstract:Drug-induced parkinsonism affects many older adults with dementia, often causing gait disturbances. New advances in vision-based human pose-estimation have opened possibilities for frequent and unobtrusive analysis of gait in residential settings. This work proposes novel spatial-temporal graph convolutional network (ST-GCN) architectures and training procedures to predict clinical scores of parkinsonism in gait from video of individuals with dementia. We propose a two-stage training approach consisting of a self-supervised pretraining stage that encourages the ST-GCN model to learn about gait patterns before predicting clinical scores in the finetuning stage. The proposed ST-GCN models are evaluated on joint trajectories extracted from video and are compared against traditional (ordinal, linear, random forest) regression models and temporal convolutional network baselines. Three 2D human pose-estimation libraries (OpenPose, Detectron, AlphaPose) and the Microsoft Kinect (2D and 3D) are used to extract joint trajectories of 4787 natural walking bouts from 53 older adults with dementia. A subset of 399 walks from 14 participants is annotated with scores of parkinsonism severity on the gait criteria of the Unified Parkinson's Disease Rating Scale (UPDRS) and the Simpson-Angus Scale (SAS). Our results demonstrate that ST-GCN models operating on 3D joint trajectories extracted from the Kinect consistently outperform all other models and feature sets. Prediction of parkinsonism scores in natural walking bouts of unseen participants remains a challenging task, with the best models achieving macro-averaged F1-scores of 0.53 +/- 0.03 and 0.40 +/- 0.02 for UPDRS-gait and SAS-gait, respectively. Pre-trained model and demo code for this work is available: https://github.com/TaatiTeam/stgcn_parkinsonism_prediction.
Unobtrusive Pain Monitoring in Older Adults with Dementia using Pairwise and Contrastive Training
Jan 08, 2021
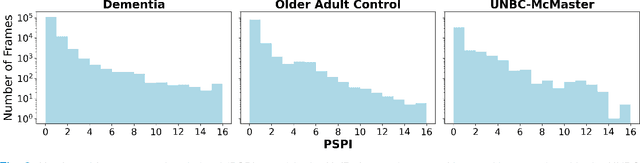
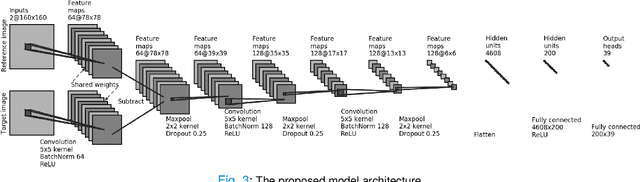
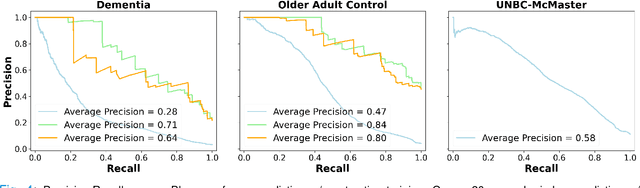
Abstract:Although pain is frequent in old age, older adults are often undertreated for pain. This is especially the case for long-term care residents with moderate to severe dementia who cannot report their pain because of cognitive impairments that accompany dementia. Nursing staff acknowledge the challenges of effectively recognizing and managing pain in long-term care facilities due to lack of human resources and, sometimes, expertise to use validated pain assessment approaches on a regular basis. Vision-based ambient monitoring will allow for frequent automated assessments so care staff could be automatically notified when signs of pain are displayed. However, existing computer vision techniques for pain detection are not validated on faces of older adults or people with dementia, and this population is not represented in existing facial expression datasets of pain. We present the first fully automated vision-based technique validated on a dementia cohort. Our contributions are threefold. First, we develop a deep learning-based computer vision system for detecting painful facial expressions on a video dataset that is collected unobtrusively from older adult participants with and without dementia. Second, we introduce a pairwise comparative inference method that calibrates to each person and is sensitive to changes in facial expression while using training data more efficiently than sequence models. Third, we introduce a fast contrastive training method that improves cross-dataset performance. Our pain estimation model outperforms baselines by a wide margin, especially when evaluated on faces of people with dementia. Pre-trained model and demo code available at https://github.com/TaatiTeam/pain_detection_demo
Estimation of Orofacial Kinematics in Parkinson's Disease: Comparison of 2D and 3D Markerless Systems for Motion Tracking
Mar 18, 2020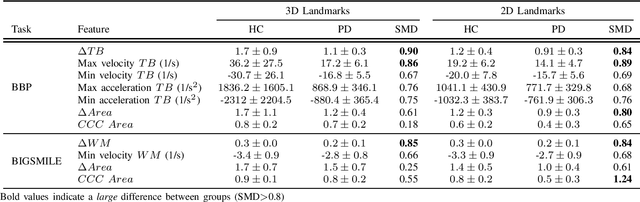
Abstract:Orofacial deficits are common in people with Parkinson's disease (PD) and their evolution might represent an important biomarker of disease progression. We are developing an automated system for assessment of orofacial function in PD that can be used in-home or in-clinic and can provide useful and objective clinical information that informs disease management. Our current approach relies on color and depth cameras for the estimation of 3D facial movements. However, depth cameras are not commonly available, might be expensive, and require specialized software for control and data processing. The objective of this paper was to evaluate if depth cameras are needed to differentiate between healthy controls and PD patients based on features extracted from orofacial kinematics. Results indicate that 2D features, extracted from color cameras only, are as informative as 3D features, extracted from color and depth cameras, differentiating healthy controls from PD patients. These results pave the way for the development of a universal system for automatic and objective assessment of orofacial function in PD.
Toward an Automatic System for Computer-Aided Assessment in Facial Palsy
Oct 25, 2019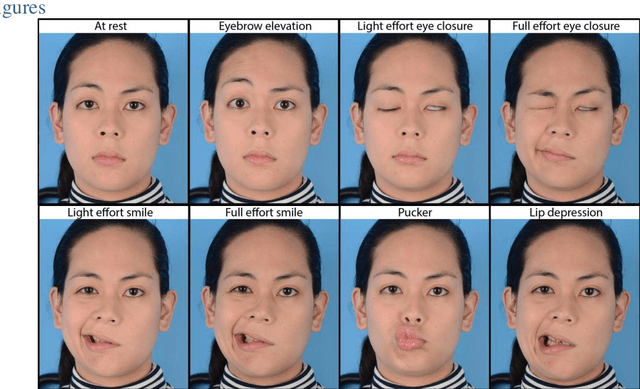
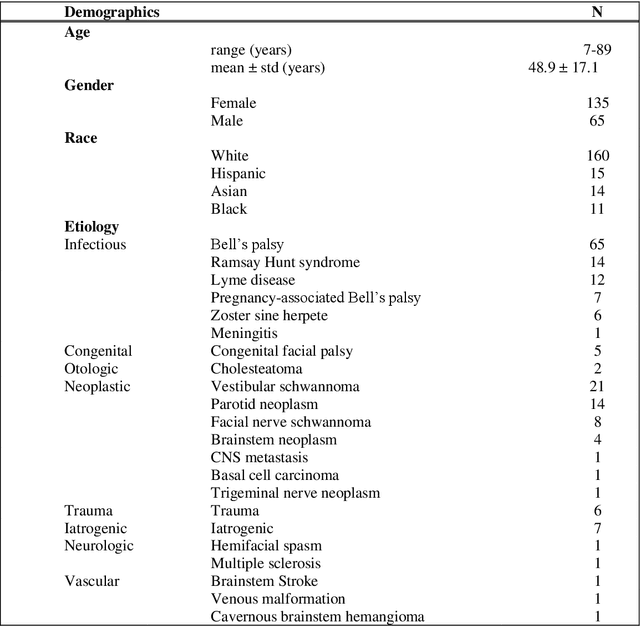
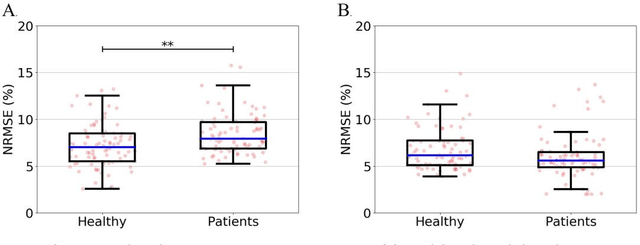
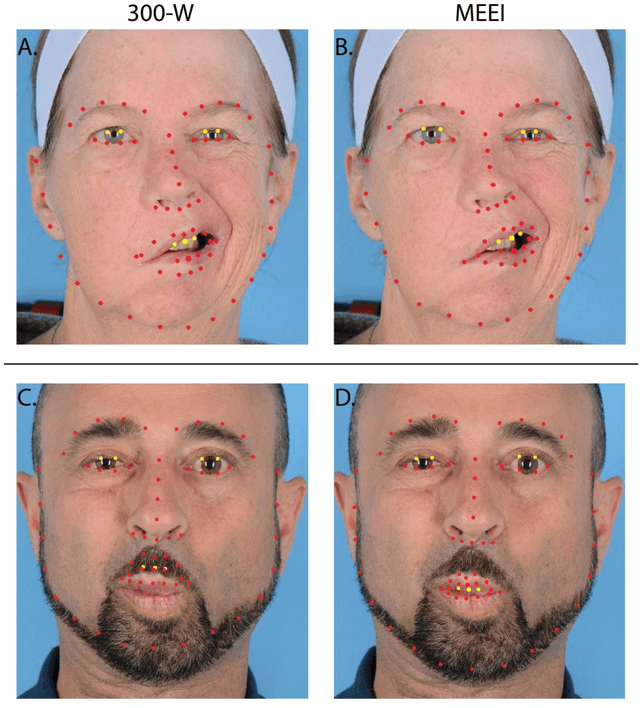
Abstract:Importance: Machine learning (ML) approaches to facial landmark localization carry great clinical potential for quantitative assessment of facial function as they enable high-throughput automated quantification of relevant facial metrics from photographs. However, translation from research settings to clinical applications requires important improvements. Objective: To develop an ML algorithm for accurate facial landmarks localization in photographs of facial palsy patients, and use it as part of an automated computer-aided diagnosis system. Design, Setting, and Participants: Facial landmarks were manually localized in portrait photographs of eight expressions obtained from 200 facial palsy patients and 10 controls. A novel ML model for automated facial landmark localization was trained using this disease-specific database. Model output was compared to manual annotations and the output of a model trained using a larger database consisting only of healthy subjects. Model accuracy was evaluated by the normalized root mean square error (NRMSE) between algorithms' prediction and manual annotations. Results: Publicly available algorithms provide poor results when applied to patients compared to healthy controls (NRMSE, 8.56 +/- 2.16 vs. 7.09 +/- 2.34, p << 0.01). We found significant improvement in facial landmark localization accuracy for the clinical population when using a model trained with a relatively small number patients' photographs (1440) compared to a model trained using several thousand more images of healthy faces (NRMSE, 6.03 +/- 2.43 vs. 8.56 +/- 2.16, p << 0.01). Conclusions: Retraining a landmark detection model with a small number of clinical images significantly improved landmark detection performance in frontal view photographs of the clinical population. These results represent the first steps towards an automatic system for computer-aided assessment in facial palsy.
Limitations and Biases in Facial Landmark Detection -- An Empirical Study on Older Adults with Dementia
May 17, 2019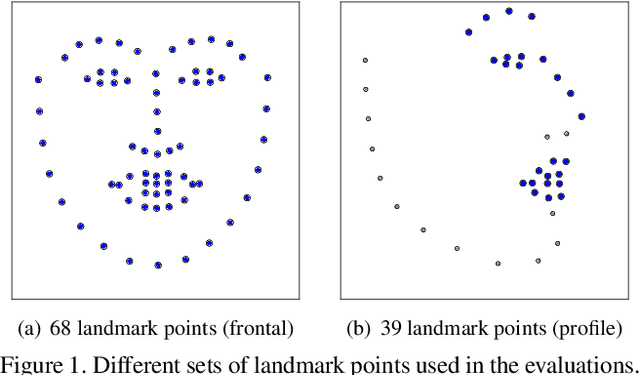
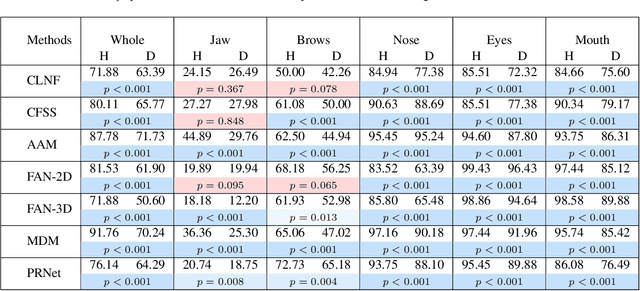
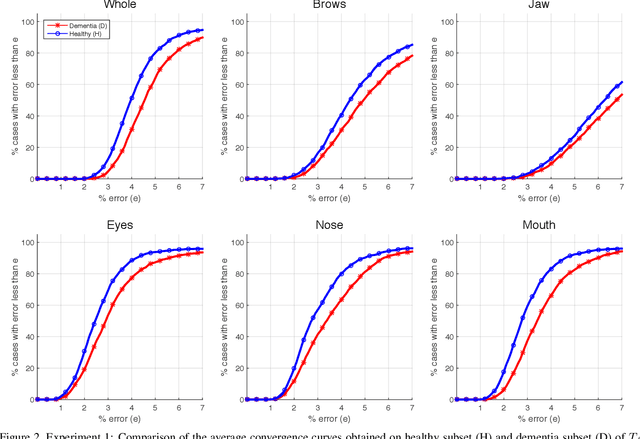
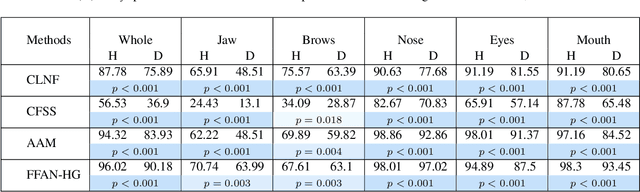
Abstract:Accurate facial expression analysis is an essential step in various clinical applications that involve physical and mental health assessments of older adults (e.g. diagnosis of pain or depression). Although remarkable progress has been achieved toward developing robust facial landmark detection methods, state-of-the-art methods still face many challenges when encountering uncontrolled environments, different ranges of facial expressions, and different demographics of the population. A recent study has revealed that the health status of individuals can also affect the performance of facial landmark detection methods on front views of faces. In this work, we investigate this matter in a much greater context using seven facial landmark detection methods. We perform our evaluation not only on frontal faces but also on profile faces and in various regions of the face. Our results shed light on limitations of the existing methods and challenges of applying these methods in clinical settings by indicating: 1) a significant difference between the performance of state-of-the-art when tested on the profile or frontal faces of individuals with vs. without dementia; 2) insights on the existing bias for all regions of the face; and 3) the presence of this bias despite re-training/fine-tuning with various configurations of six datasets.
A Hybrid Instance-based Transfer Learning Method
Dec 03, 2018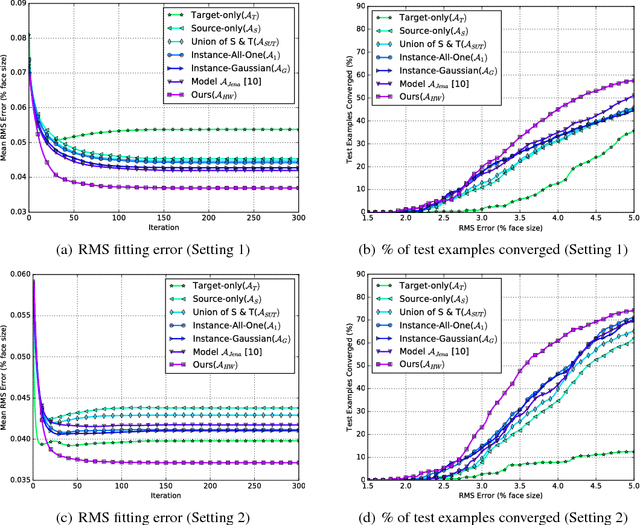
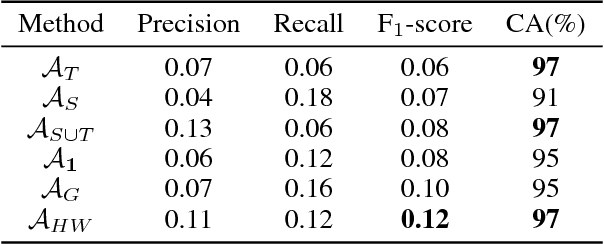
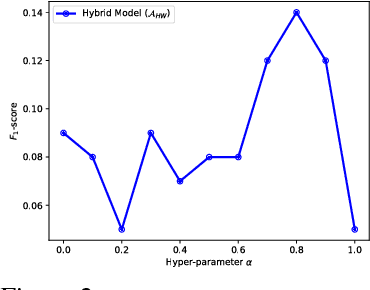

Abstract:In recent years, supervised machine learning models have demonstrated tremendous success in a variety of application domains. Despite the promising results, these successful models are data hungry and their performance relies heavily on the size of training data. However, in many healthcare applications it is difficult to collect sufficiently large training datasets. Transfer learning can help overcome this issue by transferring the knowledge from readily available datasets (source) to a new dataset (target). In this work, we propose a hybrid instance-based transfer learning method that outperforms a set of baselines including state-of-the-art instance-based transfer learning approaches. Our method uses a probabilistic weighting strategy to fuse information from the source domain to the model learned in the target domain. Our method is generic, applicable to multiple source domains, and robust with respect to negative transfer. We demonstrate the effectiveness of our approach through extensive experiments for two different applications.
Learning to Unlearn: Building Immunity to Dataset Bias in Medical Imaging Studies
Dec 03, 2018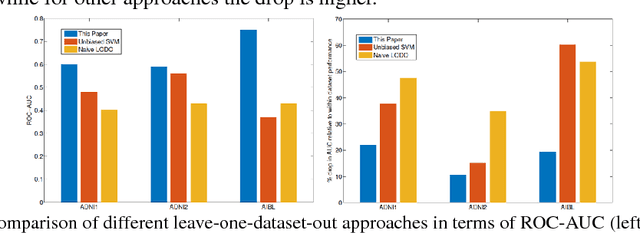
Abstract:Medical imaging machine learning algorithms are usually evaluated on a single dataset. Although training and testing are performed on different subsets of the dataset, models built on one study show limited capability to generalize to other studies. While database bias has been recognized as a serious problem in the computer vision community, it has remained largely unnoticed in medical imaging research. Transfer learning thus remains confined to the re-use of feature representations requiring re-training on the new dataset. As a result, machine learning models do not generalize even when trained on imaging datasets that were captured to study the same variable of interest. The ability to transfer knowledge gleaned from one study to another, without the need for re-training, if possible, would provide reassurance that the models are learning knowledge fundamental to the problem under study instead of latching onto the idiosyncracies of a dataset. In this paper, we situate the problem of dataset bias in the context of medical imaging studies. We show empirical evidence that such a problem exists in medical datasets. We then present a framework to unlearn study membership as a means to handle the problem of database bias. Our main idea is to take the data from the original feature space to an intermediate space where the data points are indistinguishable in terms of which study they come from, while maintaining the recognition capability with respect to the variable of interest. This will promote models which learn the more general properties of the etiology under study instead of aligning to dataset-specific peculiarities. Essentially, our proposed model learns to unlearn the dataset bias.
Subspace Selection to Suppress Confounding Source Domain Information in AAM Transfer Learning
Oct 03, 2017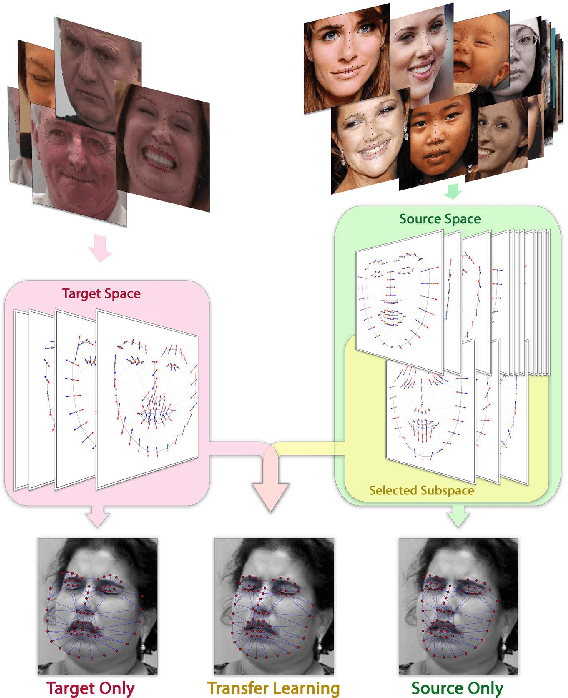
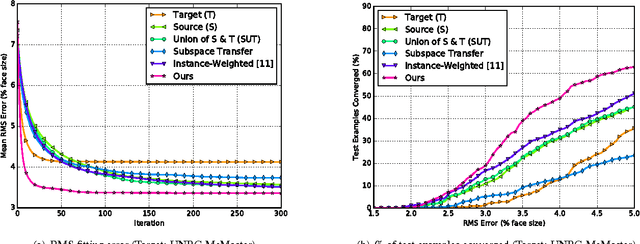
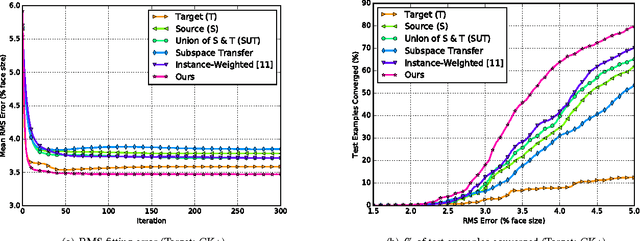
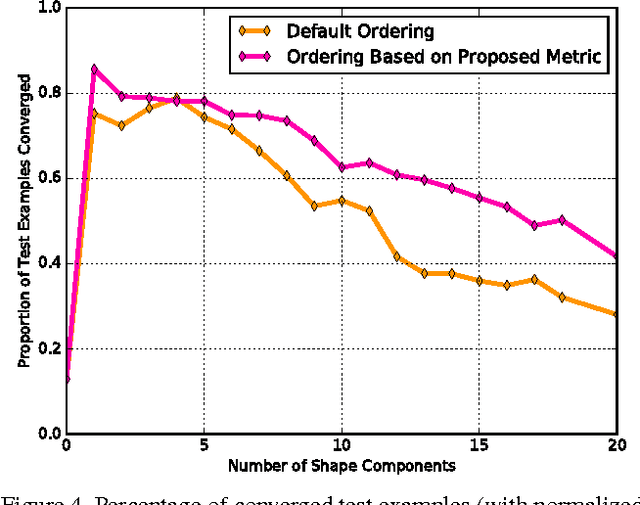
Abstract:Active appearance models (AAMs) are a class of generative models that have seen tremendous success in face analysis. However, model learning depends on the availability of detailed annotation of canonical landmark points. As a result, when accurate AAM fitting is required on a different set of variations (expression, pose, identity), a new dataset is collected and annotated. To overcome the need for time consuming data collection and annotation, transfer learning approaches have received recent attention. The goal is to transfer knowledge from previously available datasets (source) to a new dataset (target). We propose a subspace transfer learning method, in which we select a subspace from the source that best describes the target space. We propose a metric to compute the directional similarity between the source eigenvectors and the target subspace. We show an equivalence between this metric and the variance of target data when projected onto source eigenvectors. Using this equivalence, we select a subset of source principal directions that capture the variance in target data. To define our model, we augment the selected source subspace with the target subspace learned from a handful of target examples. In experiments done on six publicly available datasets, we show that our approach outperforms the state of the art in terms of the RMS fitting error as well as the percentage of test examples for which AAM fitting converges to the ground truth.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge