Artem Goncharov
Autonomous Uncertainty Quantification for Computational Point-of-care Sensors
Dec 24, 2025Abstract:Computational point-of-care (POC) sensors enable rapid, low-cost, and accessible diagnostics in emergency, remote and resource-limited areas that lack access to centralized medical facilities. These systems can utilize neural network-based algorithms to accurately infer a diagnosis from the signals generated by rapid diagnostic tests or sensors. However, neural network-based diagnostic models are subject to hallucinations and can produce erroneous predictions, posing a risk of misdiagnosis and inaccurate clinical decisions. To address this challenge, here we present an autonomous uncertainty quantification technique developed for POC diagnostics. As our testbed, we used a paper-based, computational vertical flow assay (xVFA) platform developed for rapid POC diagnosis of Lyme disease, the most prevalent tick-borne disease globally. The xVFA platform integrates a disposable paper-based assay, a handheld optical reader and a neural network-based inference algorithm, providing rapid and cost-effective Lyme disease diagnostics in under 20 min using only 20 uL of patient serum. By incorporating a Monte Carlo dropout (MCDO)-based uncertainty quantification approach into the diagnostics pipeline, we identified and excluded erroneous predictions with high uncertainty, significantly improving the sensitivity and reliability of the xVFA in an autonomous manner, without access to the ground truth diagnostic information of patients. Blinded testing using new patient samples demonstrated an increase in diagnostic sensitivity from 88.2% to 95.7%, indicating the effectiveness of MCDO-based uncertainty quantification in enhancing the robustness of neural network-driven computational POC sensing systems.
Deep learning-enhanced dual-mode multiplexed optical sensor for point-of-care diagnostics of cardiovascular diseases
Dec 24, 2025Abstract:Rapid and accessible cardiac biomarker testing is essential for the timely diagnosis and risk assessment of myocardial infarction (MI) and heart failure (HF), two interrelated conditions that frequently coexist and drive recurrent hospitalizations with high mortality. However, current laboratory and point-of-care testing systems are limited by long turnaround times, narrow dynamic ranges for the tested biomarkers, and single-analyte formats that fail to capture the complexity of cardiovascular disease. Here, we present a deep learning-enhanced dual-mode multiplexed vertical flow assay (xVFA) with a portable optical reader and a neural network-based quantification pipeline. This optical sensor integrates colorimetric and chemiluminescent detection within a single paper-based cartridge to complementarily cover a large dynamic range (spanning ~6 orders of magnitude) for both low- and high-abundance biomarkers, while maintaining quantitative accuracy. Using 50 uL of serum, the optical sensor simultaneously quantifies cardiac troponin I (cTnI), creatine kinase-MB (CK-MB), and N-terminal pro-B-type natriuretic peptide (NT-proBNP) within 23 min. The xVFA achieves sub-pg/mL sensitivity for cTnI and sub-ng/mL sensitivity for CK-MB and NT-proBNP, spanning the clinically relevant ranges for these biomarkers. Neural network models trained and blindly tested on 92 patient serum samples yielded a robust quantification performance (Pearson's r > 0.96 vs. reference assays). By combining high sensitivity, multiplexing, and automation in a compact and cost-effective optical sensor format, the dual-mode xVFA enables rapid and quantitative cardiovascular diagnostics at the point of care.
An insertable glucose sensor using a compact and cost-effective phosphorescence lifetime imager and machine learning
Jun 12, 2024Abstract:Optical continuous glucose monitoring (CGM) systems are emerging for personalized glucose management owing to their lower cost and prolonged durability compared to conventional electrochemical CGMs. Here, we report a computational CGM system, which integrates a biocompatible phosphorescence-based insertable biosensor and a custom-designed phosphorescence lifetime imager (PLI). This compact and cost-effective PLI is designed to capture phosphorescence lifetime images of an insertable sensor through the skin, where the lifetime of the emitted phosphorescence signal is modulated by the local concentration of glucose. Because this phosphorescence signal has a very long lifetime compared to tissue autofluorescence or excitation leakage processes, it completely bypasses these noise sources by measuring the sensor emission over several tens of microseconds after the excitation light is turned off. The lifetime images acquired through the skin are processed by neural network-based models for misalignment-tolerant inference of glucose levels, accurately revealing normal, low (hypoglycemia) and high (hyperglycemia) concentration ranges. Using a 1-mm thick skin phantom mimicking the optical properties of human skin, we performed in vitro testing of the PLI using glucose-spiked samples, yielding 88.8% inference accuracy, also showing resilience to random and unknown misalignments within a lateral distance of ~4.7 mm with respect to the position of the insertable sensor underneath the skin phantom. Furthermore, the PLI accurately identified larger lateral misalignments beyond 5 mm, prompting user intervention for re-alignment. The misalignment-resilient glucose concentration inference capability of this compact and cost-effective phosphorescence lifetime imager makes it an appealing wearable diagnostics tool for real-time tracking of glucose and other biomarkers.
Neural network-based on-chip spectroscopy using a scalable plasmonic encoder
Dec 01, 2020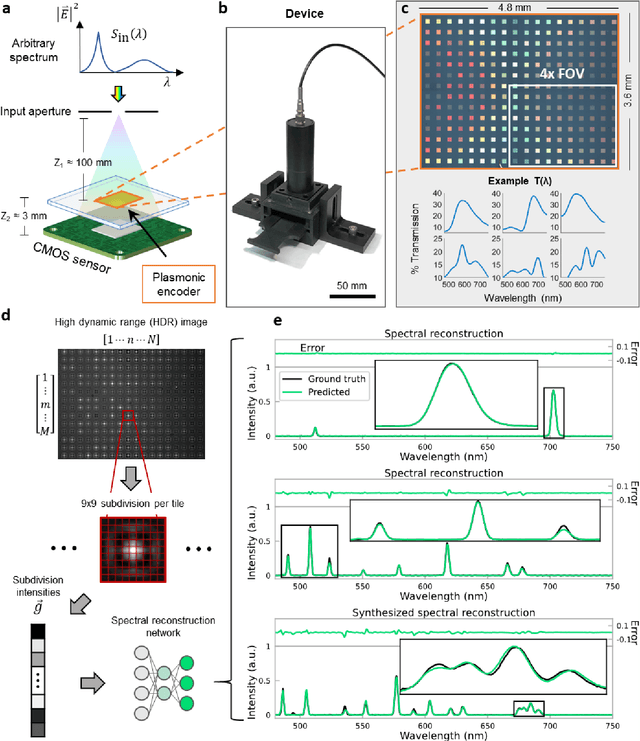
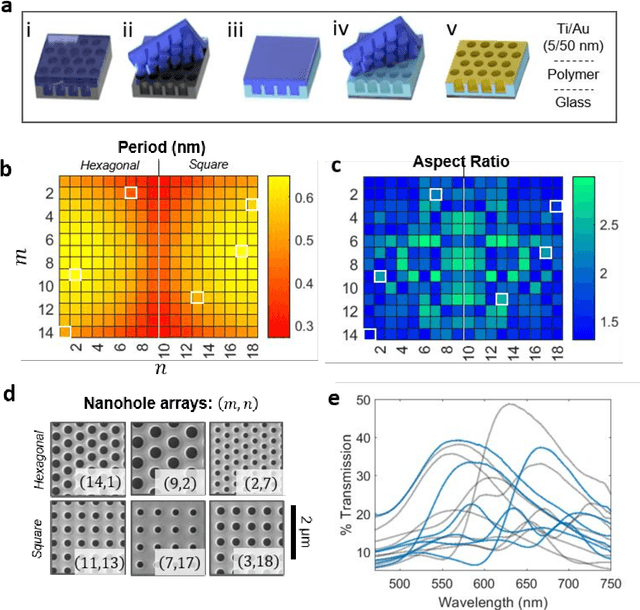
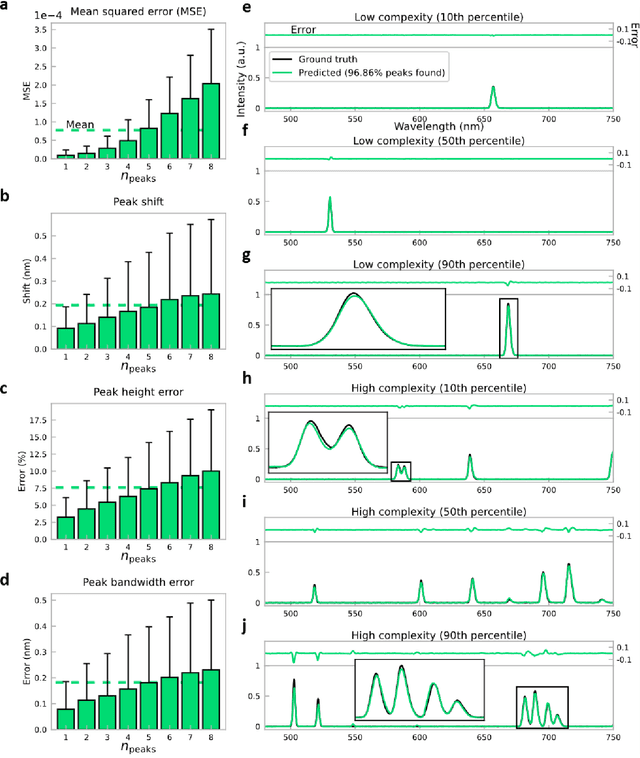
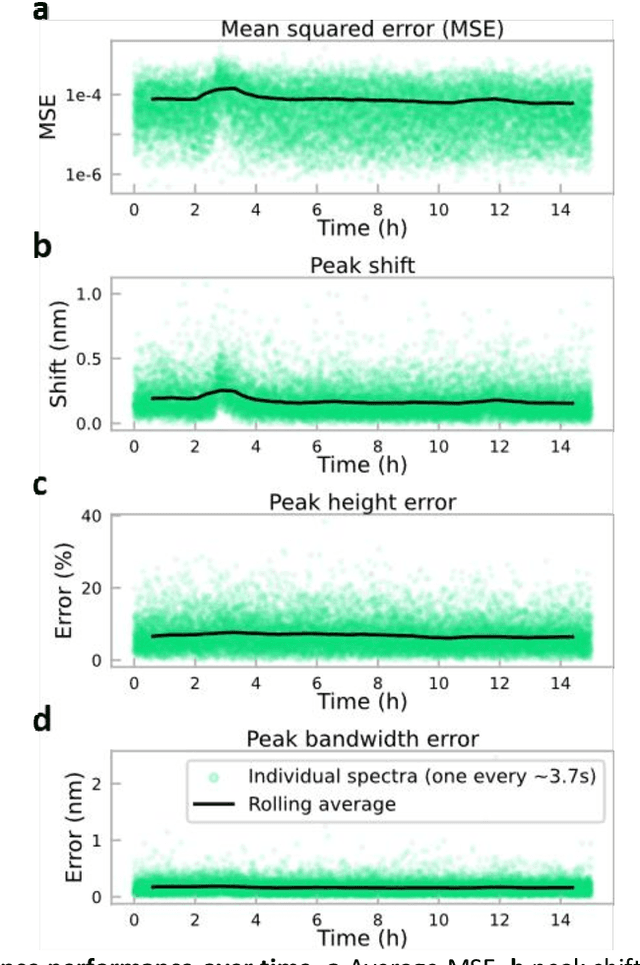
Abstract:Conventional spectrometers are limited by trade-offs set by size, cost, signal-to-noise ratio (SNR), and spectral resolution. Here, we demonstrate a deep learning-based spectral reconstruction framework, using a compact and low-cost on-chip sensing scheme that is not constrained by the design trade-offs inherent to grating-based spectroscopy. The system employs a plasmonic spectral encoder chip containing 252 different tiles of nanohole arrays fabricated using a scalable and low-cost imprint lithography method, where each tile has a unique geometry and, thus, a unique optical transmission spectrum. The illumination spectrum of interest directly impinges upon the plasmonic encoder, and a CMOS image sensor captures the transmitted light, without any lenses, gratings, or other optical components in between, making the entire hardware highly compact, light-weight and field-portable. A trained neural network then reconstructs the unknown spectrum using the transmitted intensity information from the spectral encoder in a feed-forward and non-iterative manner. Benefiting from the parallelization of neural networks, the average inference time per spectrum is ~28 microseconds, which is orders of magnitude faster compared to other computational spectroscopy approaches. When blindly tested on unseen new spectra (N = 14,648) with varying complexity, our deep-learning based system identified 96.86% of the spectral peaks with an average peak localization error, bandwidth error, and height error of 0.19 nm, 0.18 nm, and 7.60%, respectively. This system is also highly tolerant to fabrication defects that may arise during the imprint lithography process, which further makes it ideal for applications that demand cost-effective, field-portable and sensitive high-resolution spectroscopy tools.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge