Arash Mehrjou
Diffusion Models for Video Prediction and Infilling
Jun 15, 2022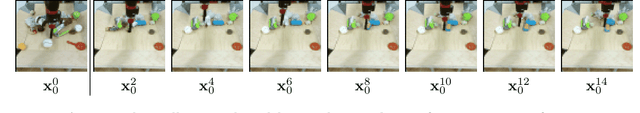


Abstract:To predict and anticipate future outcomes or reason about missing information in a sequence is a key ability for agents to be able to make intelligent decisions. This requires strong temporally coherent generative capabilities. Diffusion models have shown huge success in several generative tasks lately, but have not been extensively explored in the video domain. We present Random-Mask Video Diffusion (RaMViD), which extends image diffusion models to videos using 3D convolutions, and introduces a new conditioning technique during training. By varying the mask we condition on, the model is able to perform video prediction, infilling and upsampling. Since we do not use concatenation to condition on a mask, as done in most conditionally trained diffusion models, we are able to decrease the memory footprint. We evaluated the model on two benchmark datasets for video prediction and one for video generation on which we achieved competitive results. On Kinetics-600 we achieved state-of-the-art for video prediction.
Federated Learning in Multi-Center Critical Care Research: A Systematic Case Study using the eICU Database
Apr 20, 2022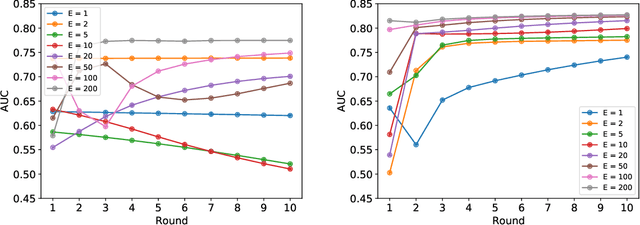
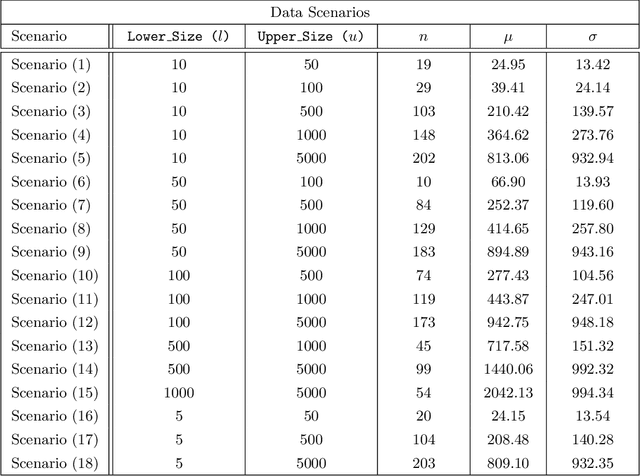
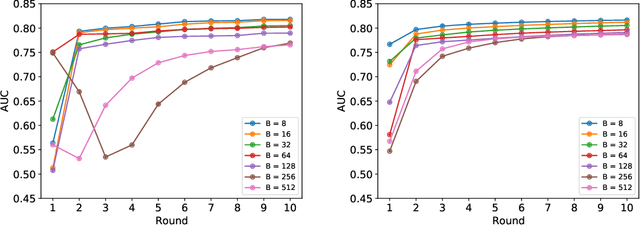
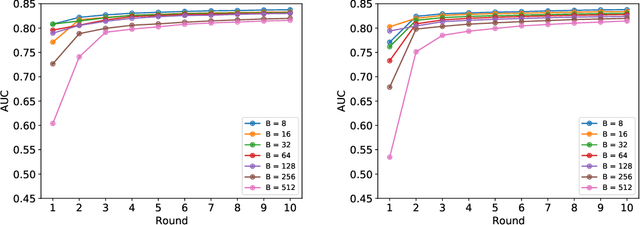
Abstract:Federated learning (FL) has been proposed as a method to train a model on different units without exchanging data. This offers great opportunities in the healthcare sector, where large datasets are available but cannot be shared to ensure patient privacy. We systematically investigate the effectiveness of FL on the publicly available eICU dataset for predicting the survival of each ICU stay. We employ Federated Averaging as the main practical algorithm for FL and show how its performance changes by altering three key hyper-parameters, taking into account that clients can significantly vary in size. We find that in many settings, a large number of local training epochs improves the performance while at the same time reducing communication costs. Furthermore, we outline in which settings it is possible to have only a low number of hospitals participating in each federated update round. When many hospitals with low patient counts are involved, the effect of overfitting can be avoided by decreasing the batchsize. This study thus contributes toward identifying suitable settings for running distributed algorithms such as FL on clinical datasets.
Physical Derivatives: Computing policy gradients by physical forward-propagation
Jan 15, 2022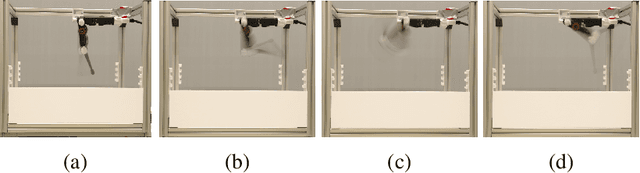
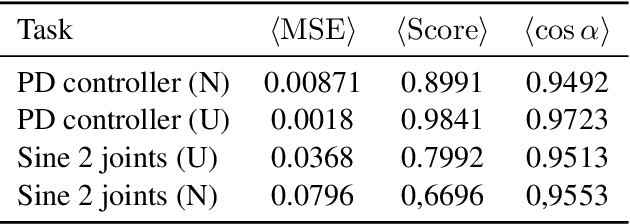
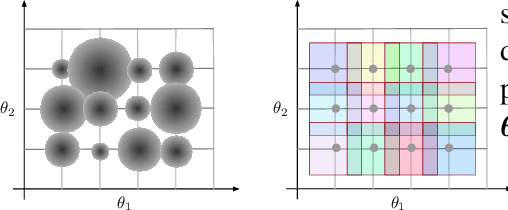
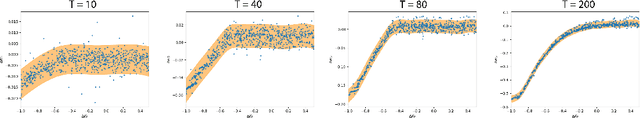
Abstract:Model-free and model-based reinforcement learning are two ends of a spectrum. Learning a good policy without a dynamic model can be prohibitively expensive. Learning the dynamic model of a system can reduce the cost of learning the policy, but it can also introduce bias if it is not accurate. We propose a middle ground where instead of the transition model, the sensitivity of the trajectories with respect to the perturbation of the parameters is learned. This allows us to predict the local behavior of the physical system around a set of nominal policies without knowing the actual model. We assay our method on a custom-built physical robot in extensive experiments and show the feasibility of the approach in practice. We investigate potential challenges when applying our method to physical systems and propose solutions to each of them.
GalilAI: Out-of-Task Distribution Detection using Causal Active Experimentation for Safe Transfer RL
Oct 29, 2021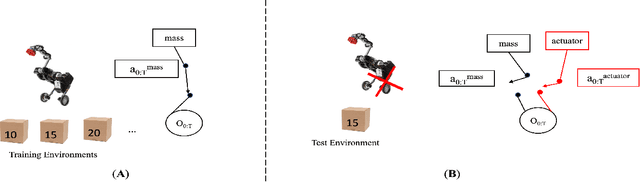
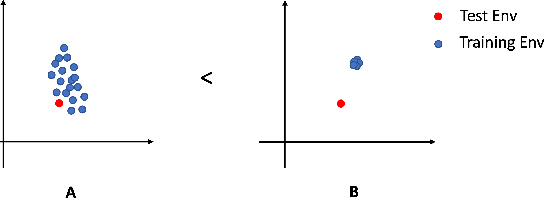
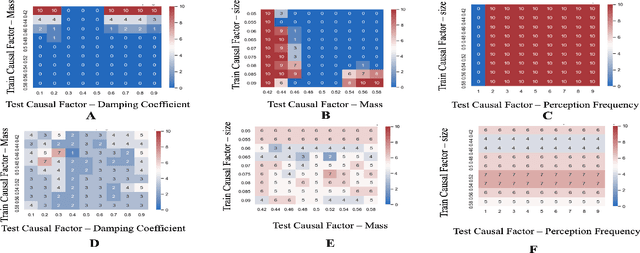
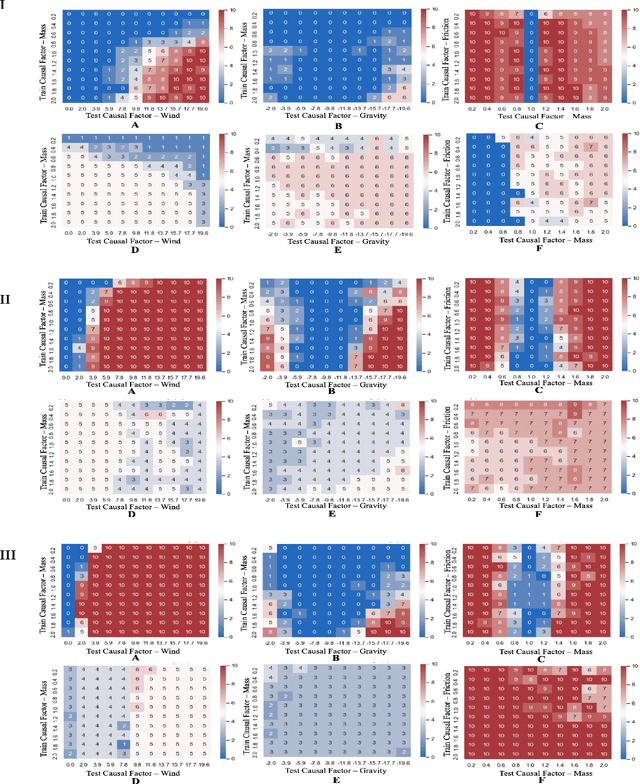
Abstract:Out-of-distribution (OOD) detection is a well-studied topic in supervised learning. Extending the successes in supervised learning methods to the reinforcement learning (RL) setting, however, is difficult due to the data generating process - RL agents actively query their environment for data, and the data are a function of the policy followed by the agent. An agent could thus neglect a shift in the environment if its policy did not lead it to explore the aspect of the environment that shifted. Therefore, to achieve safe and robust generalization in RL, there exists an unmet need for OOD detection through active experimentation. Here, we attempt to bridge this lacuna by first defining a causal framework for OOD scenarios or environments encountered by RL agents in the wild. Then, we propose a novel task: that of Out-of-Task Distribution (OOTD) detection. We introduce an RL agent that actively experiments in a test environment and subsequently concludes whether it is OOTD or not. We name our method GalilAI, in honor of Galileo Galilei, as it discovers, among other causal processes, that gravitational acceleration is independent of the mass of a body. Finally, we propose a simple probabilistic neural network baseline for comparison, which extends extant Model-Based RL. We find that GalilAI outperforms the baseline significantly. See visualizations of our method https://galil-ai.github.io/
GeneDisco: A Benchmark for Experimental Design in Drug Discovery
Oct 22, 2021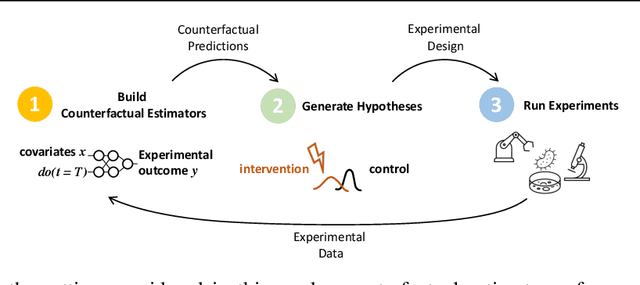
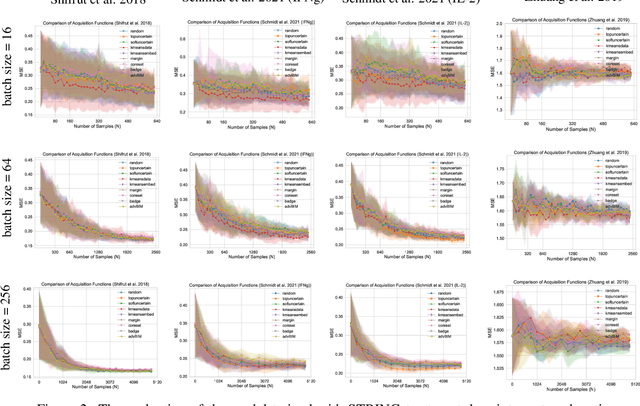
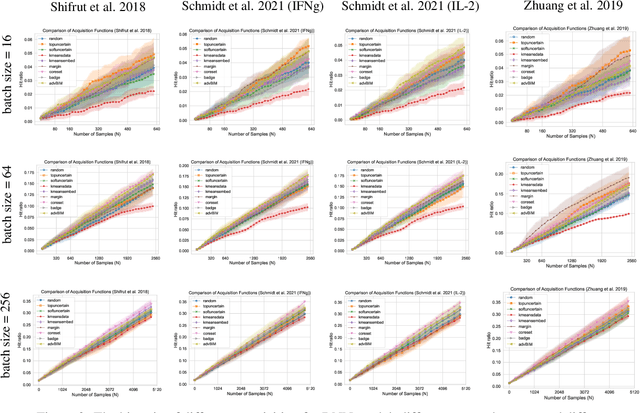
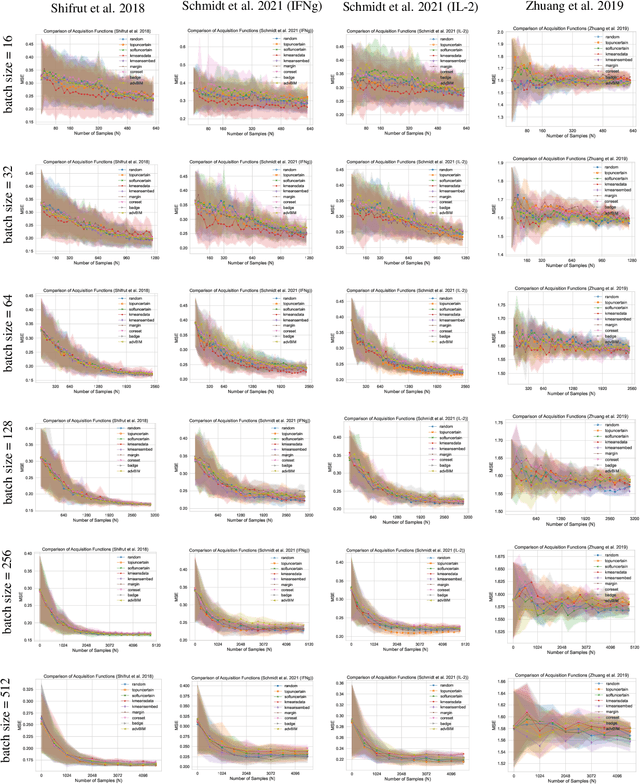
Abstract:In vitro cellular experimentation with genetic interventions, using for example CRISPR technologies, is an essential step in early-stage drug discovery and target validation that serves to assess initial hypotheses about causal associations between biological mechanisms and disease pathologies. With billions of potential hypotheses to test, the experimental design space for in vitro genetic experiments is extremely vast, and the available experimental capacity - even at the largest research institutions in the world - pales in relation to the size of this biological hypothesis space. Machine learning methods, such as active and reinforcement learning, could aid in optimally exploring the vast biological space by integrating prior knowledge from various information sources as well as extrapolating to yet unexplored areas of the experimental design space based on available data. However, there exist no standardised benchmarks and data sets for this challenging task and little research has been conducted in this area to date. Here, we introduce GeneDisco, a benchmark suite for evaluating active learning algorithms for experimental design in drug discovery. GeneDisco contains a curated set of multiple publicly available experimental data sets as well as open-source implementations of state-of-the-art active learning policies for experimental design and exploration.
Federated Learning as a Mean-Field Game
Jul 08, 2021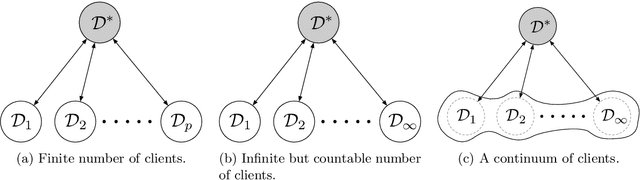
Abstract:We establish a connection between federated learning, a concept from machine learning, and mean-field games, a concept from game theory and control theory. In this analogy, the local federated learners are considered as the players and the aggregation of the gradients in a central server is the mean-field effect. We present federated learning as a differential game and discuss the properties of the equilibrium of this game. We hope this novel view to federated learning brings together researchers from these two distinct areas to work on fundamental problems of large-scale distributed and privacy-preserving learning algorithms.
Representation Learning in Continuous-Time Score-Based Generative Models
May 29, 2021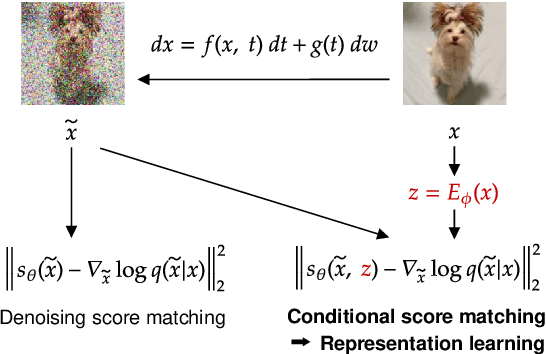
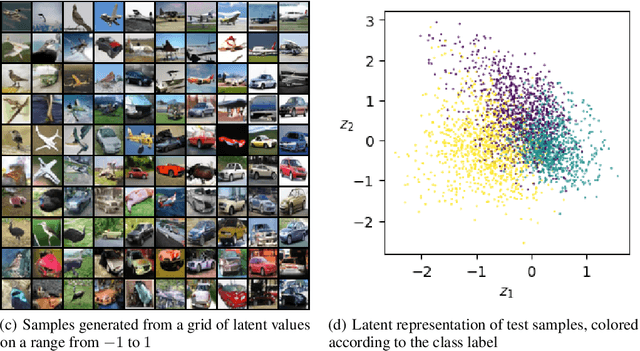
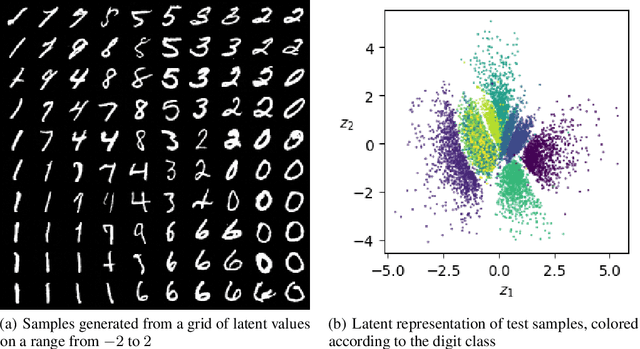
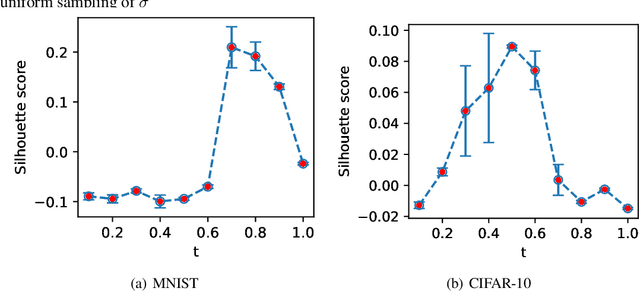
Abstract:Score-based methods represented as stochastic differential equations on a continuous time domain have recently proven successful as a non-adversarial generative model. Training such models relies on denoising score matching, which can be seen as multi-scale denoising autoencoders. Here, we augment the denoising score-matching framework to enable representation learning without any supervised signal. GANs and VAEs learn representations by directly transforming latent codes to data samples. In contrast, score-based representation learning relies on a new formulation of the denoising score-matching objective and thus encodes information needed for denoising. We show how this difference allows for manual control of the level of detail encoded in the representation.
Pyfectious: An individual-level simulator to discover optimal containment polices for epidemic diseases
Mar 24, 2021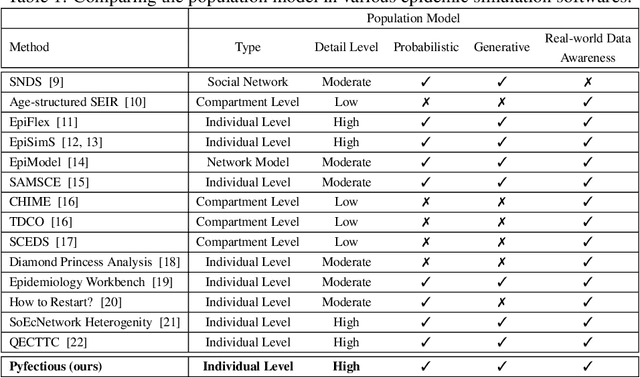
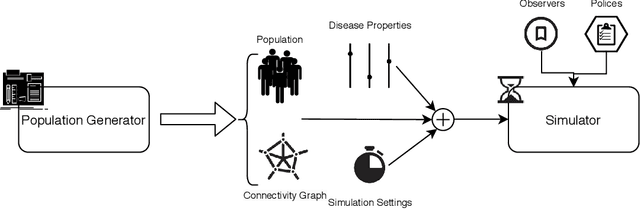
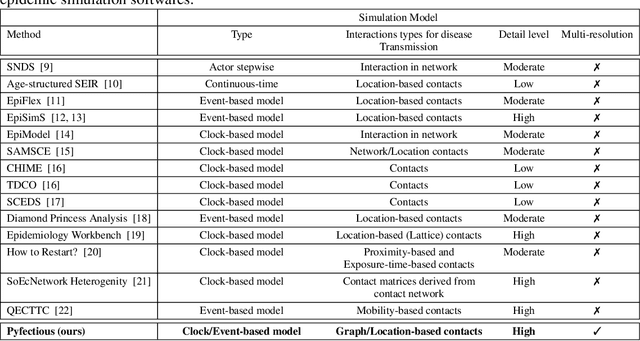
Abstract:Simulating the spread of infectious diseases in human communities is critical for predicting the trajectory of an epidemic and verifying various policies to control the devastating impacts of the outbreak. Many existing simulators are based on compartment models that divide people into a few subsets and simulate the dynamics among those subsets using hypothesized differential equations. However, these models lack the requisite granularity to study the effect of intelligent policies that influence every individual in a particular way. In this work, we introduce a simulator software capable of modeling a population structure and controlling the disease's propagation at an individualistic level. In order to estimate the confidence of the conclusions drawn from the simulator, we employ a comprehensive probabilistic approach where the entire population is constructed as a hierarchical random variable. This approach makes the inferred conclusions more robust against sampling artifacts and gives confidence bounds for decisions based on the simulation results. To showcase potential applications, the simulator parameters are set based on the formal statistics of the COVID-19 pandemic, and the outcome of a wide range of control measures is investigated. Furthermore, the simulator is used as the environment of a reinforcement learning problem to find the optimal policies to control the pandemic. The obtained experimental results indicate the simulator's adaptability and capacity in making sound predictions and a successful policy derivation example based on real-world data. As an exemplary application, our results show that the proposed policy discovery method can lead to control measures that produce significantly fewer infected individuals in the population and protect the health system against saturation.
Causal Curiosity: RL Agents Discovering Self-supervised Experiments for Causal Representation Learning
Oct 07, 2020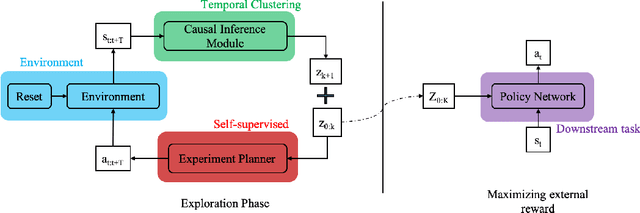
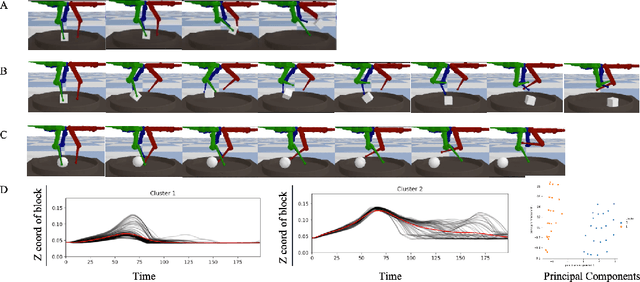
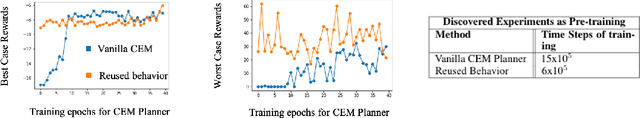
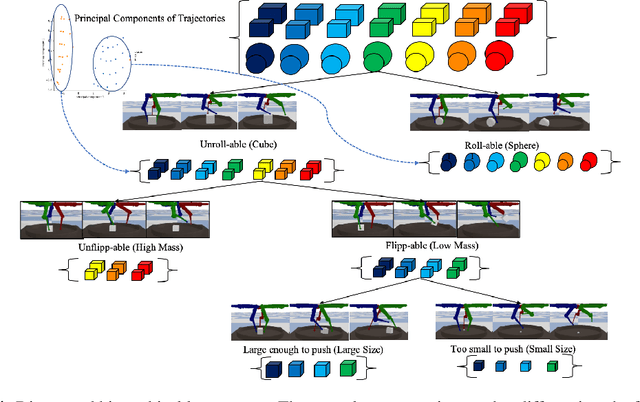
Abstract:Humans show an innate ability to learn the regularities of the world through interaction. By performing experiments in our environment, we are able to discern the causal factors of variation and infer how they affect the dynamics of our world. Analogously, here we attempt to equip reinforcement learning agents with the ability to perform experiments that facilitate a categorization of the rolled-out trajectories, and to subsequently infer the causal factors of the environment in a hierarchical manner. We introduce a novel intrinsic reward, called causal curiosity, and show that it allows our agents to learn optimal sequences of actions, and to discover causal factors in the dynamics. The learned behavior allows the agent to infer a binary quantized representation for the ground-truth causal factors in every environment. Additionally, we find that these experimental behaviors are semantically meaningful (e.g., to differentiate between heavy and light blocks, our agents learn to lift them), and are learnt in a self-supervised manner with approximately 2.5 times less data than conventional supervised planners. We show that these behaviors can be re-purposed and fine-tuned (e.g., from lifting to pushing or other downstream tasks). Finally, we show that the knowledge of causal factor representations aids zero-shot learning for more complex tasks.
Real-time Prediction of COVID-19 related Mortality using Electronic Health Records
Aug 31, 2020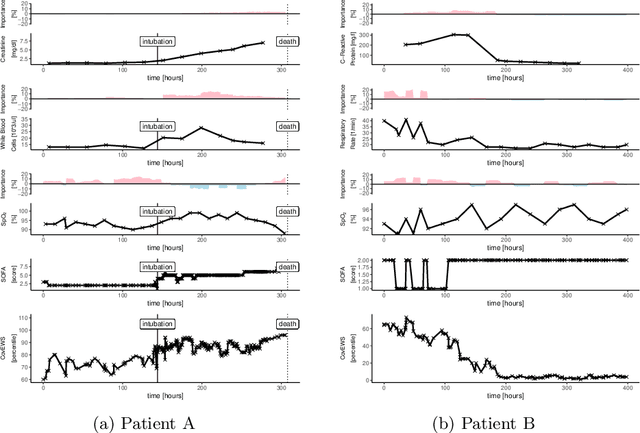
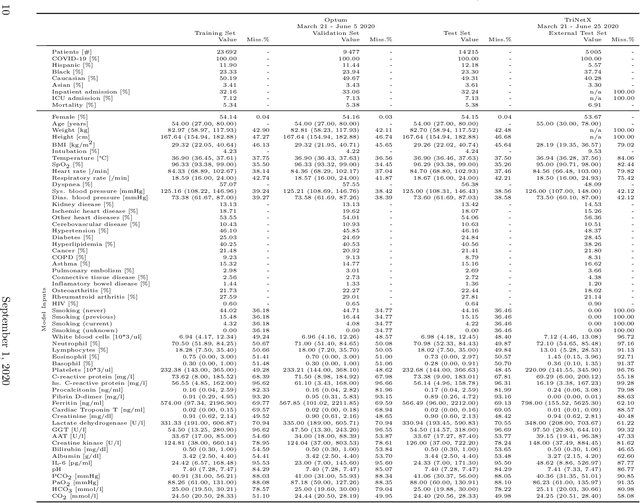
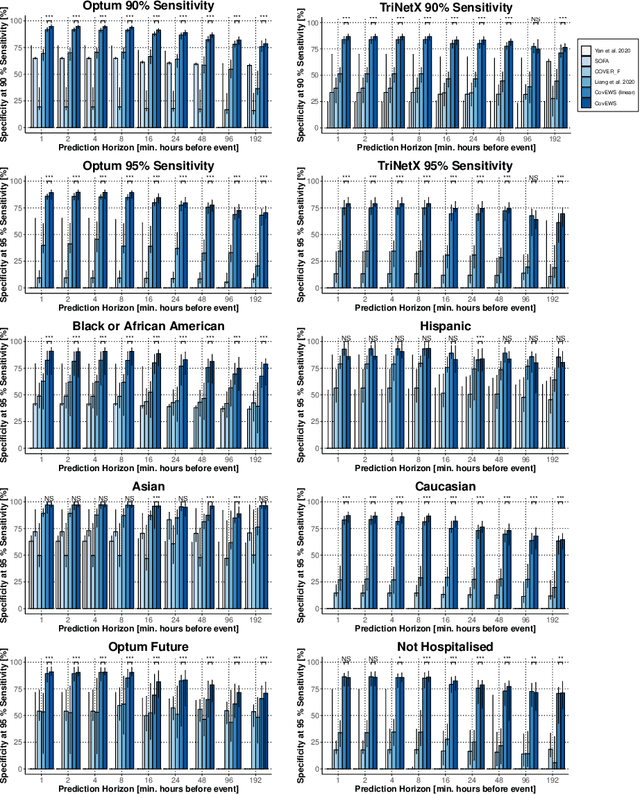
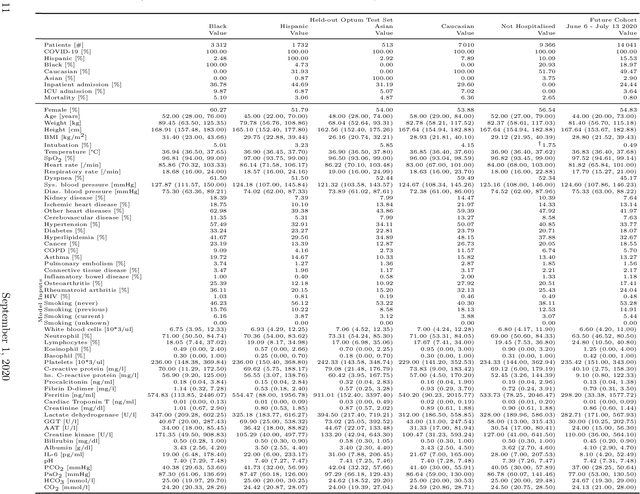
Abstract:Coronavirus Disease 2019 (COVID-19) is an emerging respiratory disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with rapid human-to-human transmission and a high case fatality rate particularly in older patients. Due to the exponential growth of infections, many healthcare systems across the world are under pressure to care for increasing amounts of at-risk patients. Given the high number of infected patients, identifying patients with the highest mortality risk early is critical to enable effective intervention and optimal prioritisation of care. Here, we present the COVID-19 Early Warning System (CovEWS), a clinical risk scoring system for assessing COVID-19 related mortality risk. CovEWS provides continuous real-time risk scores for individual patients with clinically meaningful predictive performance up to 192 hours (8 days) in advance, and is automatically derived from patients' electronic health records (EHRs) using machine learning. We trained and evaluated CovEWS using de-identified data from a cohort of 66430 COVID-19 positive patients seen at over 69 healthcare institutions in the United States (US), Australia, Malaysia and India amounting to an aggregated total of over 2863 years of patient observation time. On an external test cohort of 5005 patients, CovEWS predicts COVID-19 related mortality from $78.8\%$ ($95\%$ confidence interval [CI]: $76.0$, $84.7\%$) to $69.4\%$ ($95\%$ CI: $57.6, 75.2\%$) specificity at a sensitivity greater than $95\%$ between respectively 1 and 192 hours prior to observed mortality events - significantly outperforming existing generic and COVID-19 specific clinical risk scores. CovEWS could enable clinicians to intervene at an earlier stage, and may therefore help in preventing or mitigating COVID-19 related mortality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge