Tobias Höppe
Do Depth-Grown Models Overcome the Curse of Depth? An In-Depth Analysis
Dec 09, 2025Abstract:Gradually growing the depth of Transformers during training can not only reduce training cost but also lead to improved reasoning performance, as shown by MIDAS (Saunshi et al., 2024). Thus far, however, a mechanistic understanding of these gains has been missing. In this work, we establish a connection to recent work showing that layers in the second half of non-grown, pre-layernorm Transformers contribute much less to the final output distribution than those in the first half - also known as the Curse of Depth (Sun et al., 2025, Csordás et al., 2025). Using depth-wise analyses, we demonstrate that growth via gradual middle stacking yields more effective utilization of model depth, alters the residual stream structure, and facilitates the formation of permutable computational blocks. In addition, we propose a lightweight modification of MIDAS that yields further improvements in downstream reasoning benchmarks. Overall, this work highlights how the gradual growth of model depth can lead to the formation of distinct computational circuits and overcome the limited depth utilization seen in standard non-grown models.
Minimum-Excess-Work Guidance
May 19, 2025Abstract:We propose a regularization framework inspired by thermodynamic work for guiding pre-trained probability flow generative models (e.g., continuous normalizing flows or diffusion models) by minimizing excess work, a concept rooted in statistical mechanics and with strong conceptual connections to optimal transport. Our approach enables efficient guidance in sparse-data regimes common to scientific applications, where only limited target samples or partial density constraints are available. We introduce two strategies: Path Guidance for sampling rare transition states by concentrating probability mass on user-defined subsets, and Observable Guidance for aligning generated distributions with experimental observables while preserving entropy. We demonstrate the framework's versatility on a coarse-grained protein model, guiding it to sample transition configurations between folded/unfolded states and correct systematic biases using experimental data. The method bridges thermodynamic principles with modern generative architectures, offering a principled, efficient, and physics-inspired alternative to standard fine-tuning in data-scarce domains. Empirical results highlight improved sample efficiency and bias reduction, underscoring its applicability to molecular simulations and beyond.
In-silico biological discovery with large perturbation models
Mar 30, 2025



Abstract:Data generated in perturbation experiments link perturbations to the changes they elicit and therefore contain information relevant to numerous biological discovery tasks -- from understanding the relationships between biological entities to developing therapeutics. However, these data encompass diverse perturbations and readouts, and the complex dependence of experimental outcomes on their biological context makes it challenging to integrate insights across experiments. Here, we present the Large Perturbation Model (LPM), a deep-learning model that integrates multiple, heterogeneous perturbation experiments by representing perturbation, readout, and context as disentangled dimensions. LPM outperforms existing methods across multiple biological discovery tasks, including in predicting post-perturbation transcriptomes of unseen experiments, identifying shared molecular mechanisms of action between chemical and genetic perturbations, and facilitating the inference of gene-gene interaction networks.
Diffusion Models for Video Prediction and Infilling
Jun 15, 2022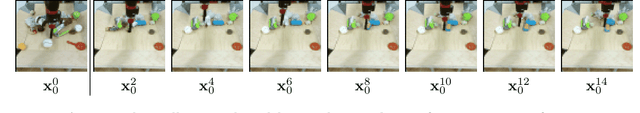
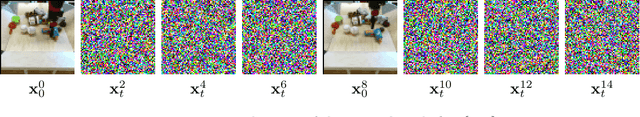


Abstract:To predict and anticipate future outcomes or reason about missing information in a sequence is a key ability for agents to be able to make intelligent decisions. This requires strong temporally coherent generative capabilities. Diffusion models have shown huge success in several generative tasks lately, but have not been extensively explored in the video domain. We present Random-Mask Video Diffusion (RaMViD), which extends image diffusion models to videos using 3D convolutions, and introduces a new conditioning technique during training. By varying the mask we condition on, the model is able to perform video prediction, infilling and upsampling. Since we do not use concatenation to condition on a mask, as done in most conditionally trained diffusion models, we are able to decrease the memory footprint. We evaluated the model on two benchmark datasets for video prediction and one for video generation on which we achieved competitive results. On Kinetics-600 we achieved state-of-the-art for video prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge