Andriy Mulyar
CoRNStack: High-Quality Contrastive Data for Better Code Ranking
Dec 01, 2024



Abstract:Effective code retrieval plays a crucial role in advancing code generation, bug fixing, and software maintenance, particularly as software systems increase in complexity. While current code embedding models have demonstrated promise in retrieving code snippets for small-scale, well-defined tasks, they often underperform in more demanding real-world applications such as bug localization within GitHub repositories. We hypothesize that a key issue is their reliance on noisy and inconsistent datasets for training, which impedes their ability to generalize to more complex retrieval scenarios. To address these limitations, we introduce CoRNStack, a large-scale, high-quality contrastive training dataset for code that spans multiple programming languages. This dataset is curated using consistency filtering to eliminate noisy positives and is further enriched with mined hard negatives, thereby facilitating more effective learning. We demonstrate that contrastive training of embedding models using CoRNStack leads to state-of-the-art performance across a variety of code retrieval tasks. Furthermore, the dataset can be leveraged for training code reranking models, a largely underexplored area compared to text reranking. Our finetuned code reranking model significantly improves the ranking quality over the retrieved results. Finally, by employing our code retriever and reranker together, we demonstrate significant improvements in function localization for GitHub issues, an important component of real-world software development.
Nomic Embed: Training a Reproducible Long Context Text Embedder
Feb 02, 2024



Abstract:This technical report describes the training of nomic-embed-text-v1, the first fully reproducible, open-source, open-weights, open-data, 8192 context length English text embedding model that outperforms both OpenAI Ada-002 and OpenAI text-embedding-3-small on short and long-context tasks. We release the training code and model weights under an Apache 2 license. In contrast with other open-source models, we release a training data loader with 235 million curated text pairs that allows for the full replication of nomic-embed-text-v1. You can find code and data to replicate the model at https://github.com/nomic-ai/contrastors
GPT4All: An Ecosystem of Open Source Compressed Language Models
Nov 06, 2023Abstract:Large language models (LLMs) have recently achieved human-level performance on a range of professional and academic benchmarks. The accessibility of these models has lagged behind their performance. State-of-the-art LLMs require costly infrastructure; are only accessible via rate-limited, geo-locked, and censored web interfaces; and lack publicly available code and technical reports. In this paper, we tell the story of GPT4All, a popular open source repository that aims to democratize access to LLMs. We outline the technical details of the original GPT4All model family, as well as the evolution of the GPT4All project from a single model into a fully fledged open source ecosystem. It is our hope that this paper acts as both a technical overview of the original GPT4All models as well as a case study on the subsequent growth of the GPT4All open source ecosystem.
MT-Clinical BERT: Scaling Clinical Information Extraction with Multitask Learning
Apr 21, 2020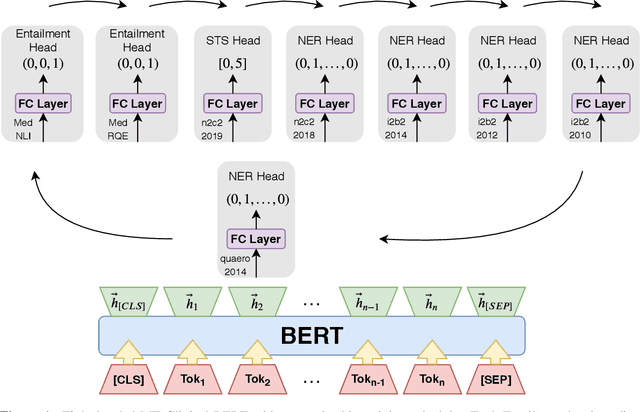
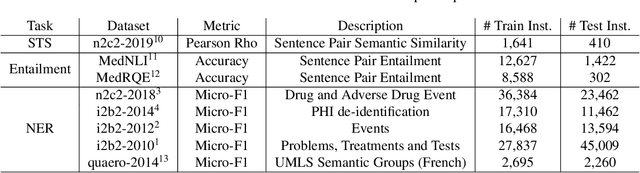
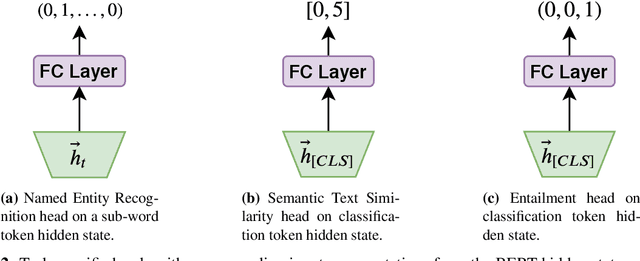
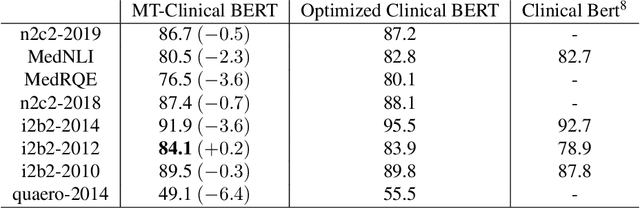
Abstract:Clinical notes contain an abundance of important but not-readily accessible information about patients. Systems to automatically extract this information rely on large amounts of training data for which their exists limited resources to create. Furthermore, they are developed dis-jointly; meaning that no information can be shared amongst task-specific systems. This bottle-neck unnecessarily complicates practical application, reduces the performance capabilities of each individual solution and associates the engineering debt of managing multiple information extraction systems. We address these challenges by developing Multitask-Clinical BERT: a single deep learning model that simultaneously performs eight clinical tasks spanning entity extraction, PHI identification, language entailment and similarity by sharing representations amongst tasks. We find our single system performs competitively with all state-the-art task-specific systems while also benefiting from massive computational benefits at inference.
Phenotyping of Clinical Notes with Improved Document Classification Models Using Contextualized Neural Language Models
Oct 30, 2019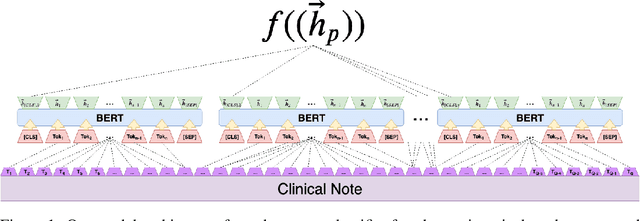
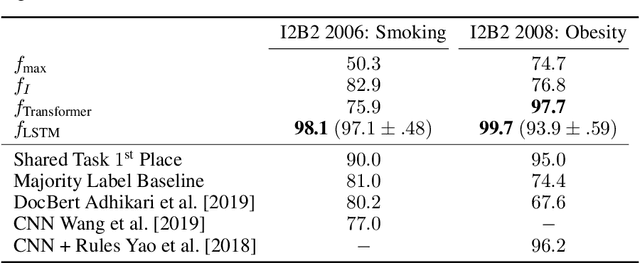
Abstract:Clinical notes contain an extensive record of a patient's health status, such as smoking status or the presence of heart conditions. However, this detail is not replicated within the structured data of electronic health systems. Phenotyping, the extraction of patient conditions from free clinical text, is a critical task which supports avariety of downstream applications such as decision support and secondary use ofmedical records. Previous work has resulted in systems which are high performing but require hand engineering, often of rules. Recent work in pretrained contextualized language models have enabled advances in representing text for a variety of tasks. We therefore explore several architectures for modeling pheno-typing that rely solely on BERT representations of the clinical note, removing the need for manual engineering. We find these architectures are competitive with or outperform existing state of the art methods on two phenotyping tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge