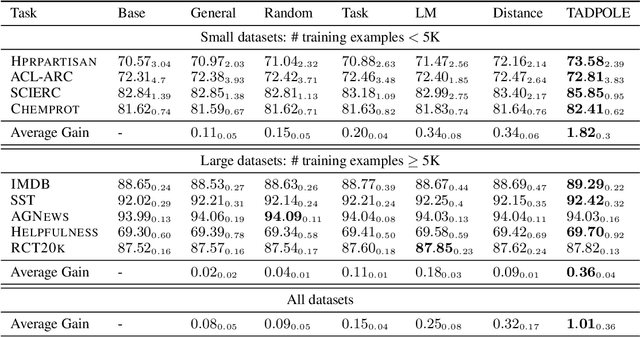
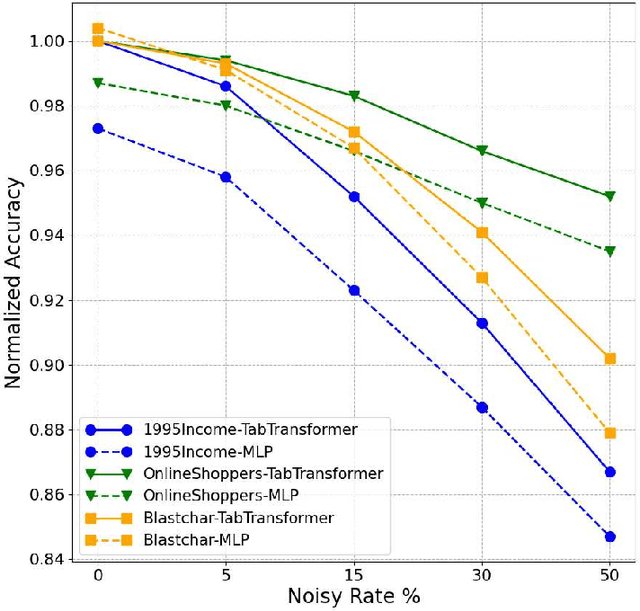
Alex Ratner
Proceedings of the First Workshop on Weakly Supervised Learning
Jul 08, 2021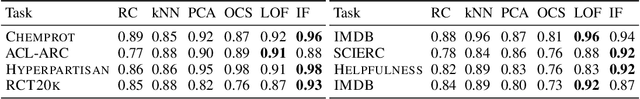

Abstract:Welcome to WeaSuL 2021, the First Workshop on Weakly Supervised Learning, co-located with ICLR 2021. In this workshop, we want to advance theory, methods and tools for allowing experts to express prior coded knowledge for automatic data annotations that can be used to train arbitrary deep neural networks for prediction. The ICLR 2021 Workshop on Weak Supervision aims at advancing methods that help modern machine-learning methods to generalize from knowledge provided by experts, in interaction with observable (unlabeled) data. In total, 15 papers were accepted. All the accepted contributions are listed in these Proceedings.
SwellShark: A Generative Model for Biomedical Named Entity Recognition without Labeled Data
Apr 20, 2017
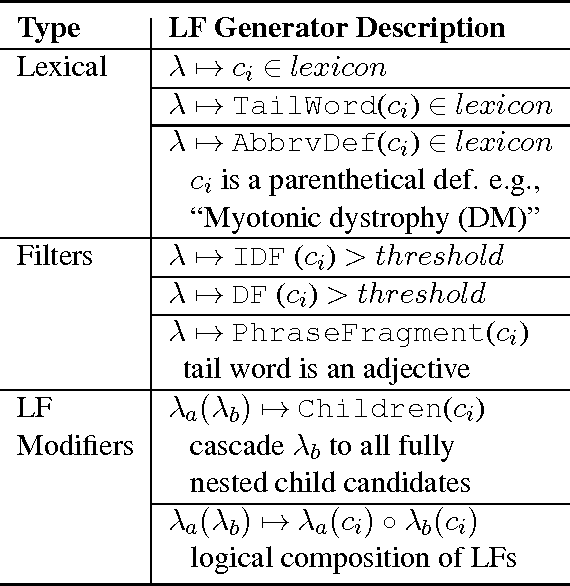
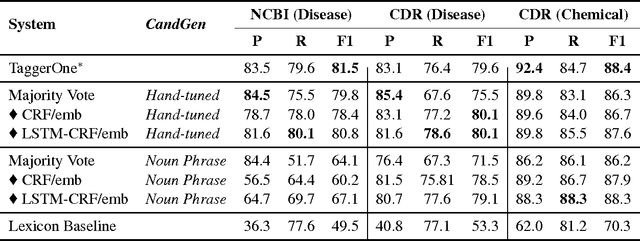
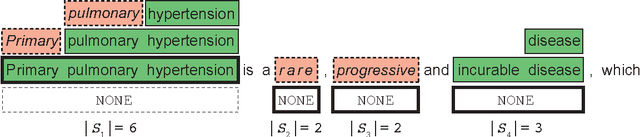
Abstract:We present SwellShark, a framework for building biomedical named entity recognition (NER) systems quickly and without hand-labeled data. Our approach views biomedical resources like lexicons as function primitives for autogenerating weak supervision. We then use a generative model to unify and denoise this supervision and construct large-scale, probabilistically labeled datasets for training high-accuracy NER taggers. In three biomedical NER tasks, SwellShark achieves competitive scores with state-of-the-art supervised benchmarks using no hand-labeled training data. In a drug name extraction task using patient medical records, one domain expert using SwellShark achieved within 5.1% of a crowdsourced annotation approach -- which originally utilized 20 teams over the course of several weeks -- in 24 hours.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge