Alan Wang
Building Performance Simulations Can Inform IoT Privacy Leaks in Buildings
Mar 26, 2023Abstract:As IoT devices become cheaper, smaller, and more ubiquitously deployed, they can reveal more information than their intended design and threaten user privacy. Indoor Environmental Quality (IEQ) sensors previously installed for energy savings and indoor health monitoring have emerged as an avenue to infer sensitive occupant information. For example, light sensors are a known conduit for inspecting room occupancy status with motion-sensitive lights. Light signals can also infer sensitive data such as occupant identity and digital screen information. To limit sensor overreach, we explore the selection of sensor placements as a methodology. Specifically, in this proof-of-concept exploration, we demonstrate the potential of physics-based simulation models to quantify the minimal number of positions necessary to capture sensitive inferences. We show how a single well-placed sensor can be sufficient in specific building contexts to holistically capture its environmental states and how additional well-placed sensors can contribute to more granular inferences. We contribute a device-agnostic and building-adaptive workflow to respectfully capture inferable occupant activity and elaborate on the implications of incorporating building simulations into sensing schemes in the real world.
Neural Pre-Processing: A Learning Framework for End-to-end Brain MRI Pre-processing
Mar 21, 2023



Abstract:Head MRI pre-processing involves converting raw images to an intensity-normalized, skull-stripped brain in a standard coordinate space. In this paper, we propose an end-to-end weakly supervised learning approach, called Neural Pre-processing (NPP), for solving all three sub-tasks simultaneously via a neural network, trained on a large dataset without individual sub-task supervision. Because the overall objective is highly under-constrained, we explicitly disentangle geometric-preserving intensity mapping (skull-stripping and intensity normalization) and spatial transformation (spatial normalization). Quantitative results show that our model outperforms state-of-the-art methods which tackle only a single sub-task. Our ablation experiments demonstrate the importance of the architecture design we chose for NPP. Furthermore, NPP affords the user the flexibility to control each of these tasks at inference time. The code and model are freely-available at \url{https://github.com/Novestars/Neural-Pre-processing}.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021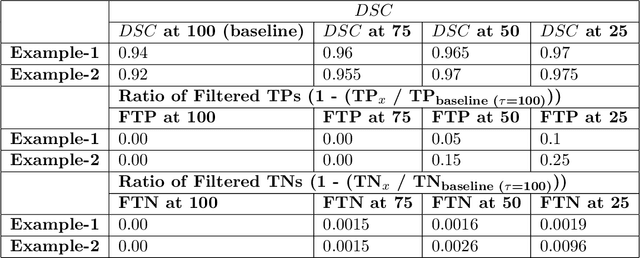
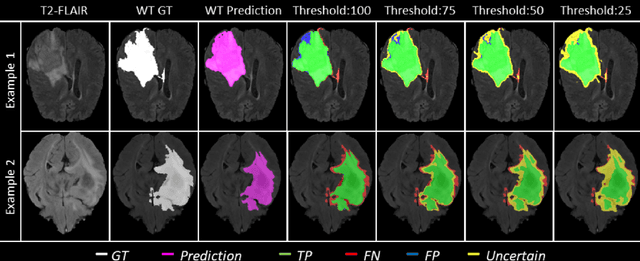
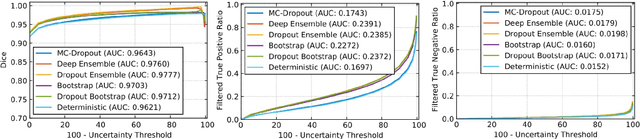
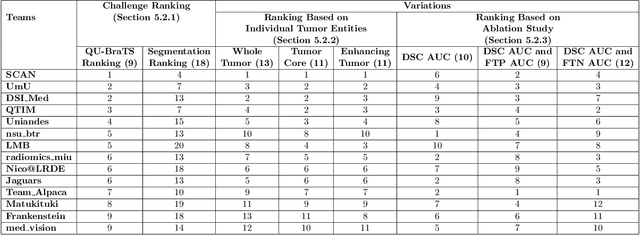
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Extending LOUPE for K-space Under-sampling Pattern Optimization in Multi-coil MRI
Jul 28, 2020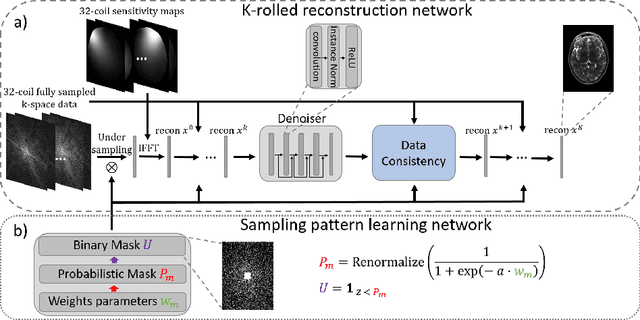
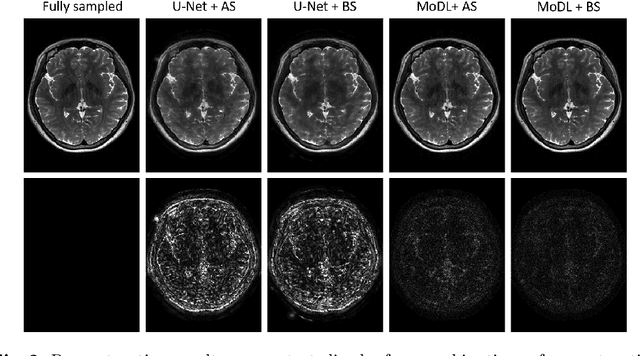
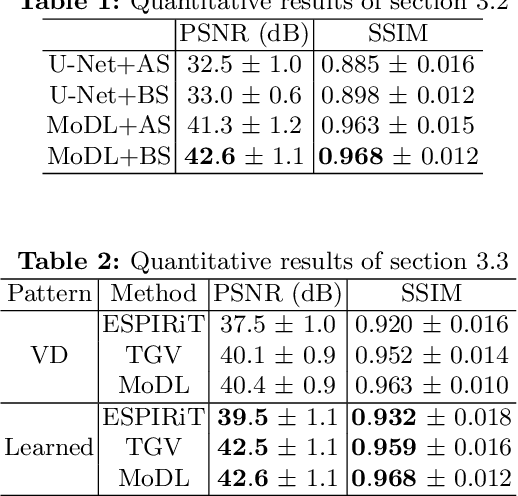
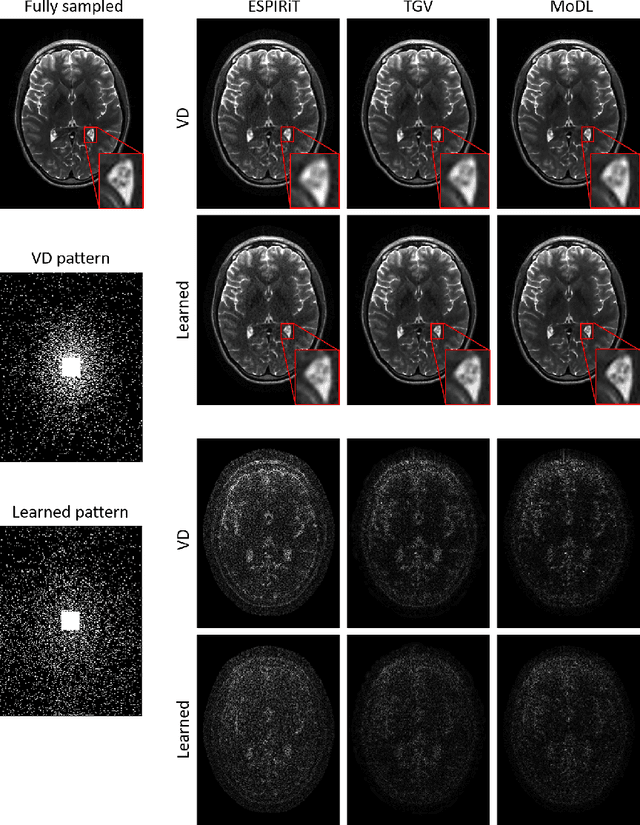
Abstract:The previously established LOUPE (Learning-based Optimization of the Under-sampling Pattern) framework for optimizing the k-space sampling pattern in MRI was extended in three folds: firstly, fully sampled multi-coil k-space data from the scanner, rather than simulated k-space data from magnitude MR images in LOUPE, was retrospectively under-sampled to optimize the under-sampling pattern of in-vivo k-space data; secondly, binary stochastic k-space sampling, rather than approximate stochastic k-space sampling of LOUPE during training, was applied together with a straight-through (ST) estimator to estimate the gradient of the threshold operation in a neural network; thirdly, modified unrolled optimization network, rather than modified U-Net in LOUPE, was used as the reconstruction network in order to reconstruct multi-coil data properly and reduce the dependency on training data. Experimental results show that when dealing with the in-vivo k-space data, unrolled optimization network with binary under-sampling block and ST estimator had better reconstruction performance compared to the ones with either U-Net reconstruction network or approximate sampling pattern optimization network, and once trained, the learned optimal sampling pattern worked better than the hand-crafted variable density sampling pattern when deployed with other conventional reconstruction methods.
4DFlowNet: Super-Resolution 4D Flow MRI using Deep Learning and Computational Fluid Dynamics
Apr 15, 2020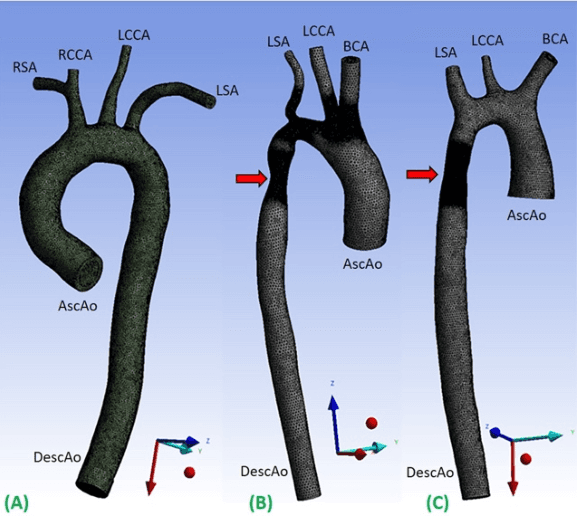
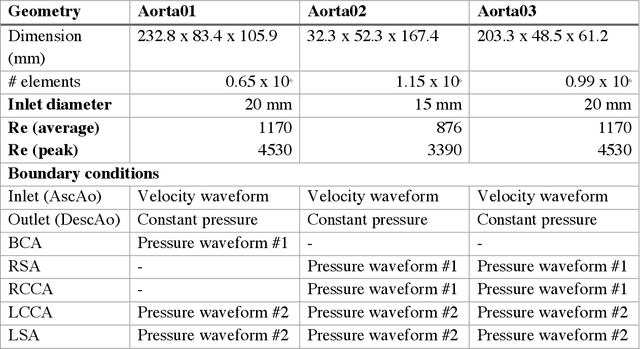
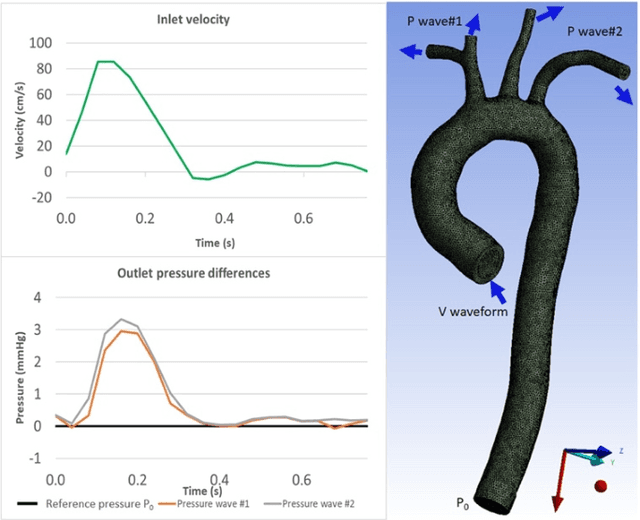
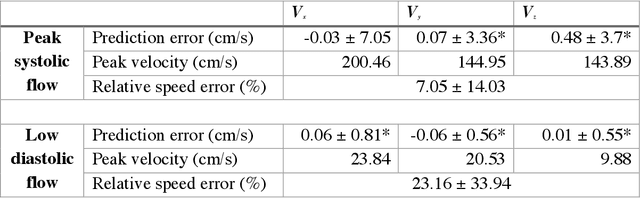
Abstract:4D-flow magnetic resonance imaging (MRI) is an emerging imaging technique where spatiotemporal 3D blood velocity can be captured with full volumetric coverage in a single non-invasive examination. This enables qualitative and quantitative analysis of hemodynamic flow parameters of the heart and great vessels. An increase in the image resolution would provide more accuracy and allow better assessment of the blood flow, especially for patients with abnormal flows. However, this must be balanced with increasing imaging time. The recent success of deep learning in generating super resolution images shows promise for implementation in medical images. We utilized computational fluid dynamics simulations to generate fluid flow simulations and represent them as synthetic 4D flow MRI data. We built our training dataset to mimic actual 4D flow MRI data with its corresponding noise distribution. Our novel 4DFlowNet network was trained on this synthetic 4D flow data and was capable in producing noise-free super resolution 4D flow phase images with upsample factor of 2. We also tested the 4DFlowNet in actual 4D flow MR images of a phantom and normal volunteer data, and demonstrated comparable results with the actual flow rate measurements giving an absolute relative error of 0.6 to 5.8% and 1.1 to 3.8% in the phantom data and normal volunteer data, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge