Isic 2020 Challenge Dataset
Papers and Code
On the Role of Calibration in Benchmarking Algorithmic Fairness for Skin Cancer Detection
Nov 10, 2025Artificial Intelligence (AI) models have demonstrated expert-level performance in melanoma detection, yet their clinical adoption is hindered by performance disparities across demographic subgroups such as gender, race, and age. Previous efforts to benchmark the performance of AI models have primarily focused on assessing model performance using group fairness metrics that rely on the Area Under the Receiver Operating Characteristic curve (AUROC), which does not provide insights into a model's ability to provide accurate estimates. In line with clinical assessments, this paper addresses this gap by incorporating calibration as a complementary benchmarking metric to AUROC-based fairness metrics. Calibration evaluates the alignment between predicted probabilities and observed event rates, offering deeper insights into subgroup biases. We assess the performance of the leading skin cancer detection algorithm of the ISIC 2020 Challenge on the ISIC 2020 Challenge dataset and the PROVE-AI dataset, and compare it with the second and third place models, focusing on subgroups defined by sex, race (Fitzpatrick Skin Tone), and age. Our findings reveal that while existing models enhance discriminative accuracy, they often over-diagnose risk and exhibit calibration issues when applied to new datasets. This study underscores the necessity for comprehensive model auditing strategies and extensive metadata collection to achieve equitable AI-driven healthcare solutions. All code is publicly available at https://github.com/bdominique/testing_strong_calibration.
* 19 pages, 4 figures. Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2025:027
MAPUNetR: A Hybrid Vision Transformer and U-Net Architecture for Efficient and Interpretable Medical Image Segmentation
Oct 29, 2024
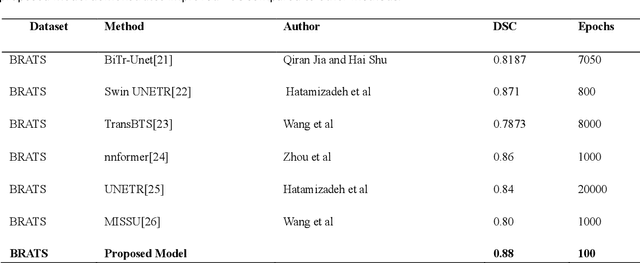
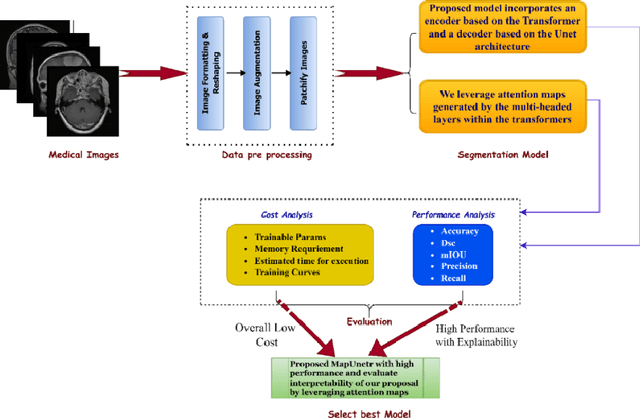
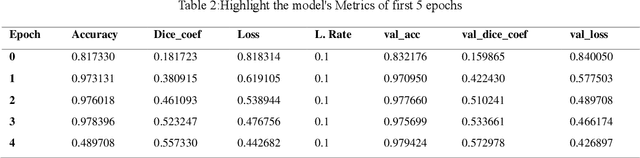
Medical image segmentation is pivotal in healthcare, enhancing diagnostic accuracy, informing treatment strategies, and tracking disease progression. This process allows clinicians to extract critical information from visual data, enabling personalized patient care. However, developing neural networks for segmentation remains challenging, especially when preserving image resolution, which is essential in detecting subtle details that influence diagnoses. Moreover, the lack of transparency in these deep learning models has slowed their adoption in clinical practice. Efforts in model interpretability are increasingly focused on making these models' decision-making processes more transparent. In this paper, we introduce MAPUNetR, a novel architecture that synergizes the strengths of transformer models with the proven U-Net framework for medical image segmentation. Our model addresses the resolution preservation challenge and incorporates attention maps highlighting segmented regions, increasing accuracy and interpretability. Evaluated on the BraTS 2020 dataset, MAPUNetR achieved a dice score of 0.88 and a dice coefficient of 0.92 on the ISIC 2018 dataset. Our experiments show that the model maintains stable performance and potential as a powerful tool for medical image segmentation in clinical practice.
Entropy-Aware Similarity for Balanced Clustering: A Case Study with Melanoma Detection
May 11, 2023Clustering data is an unsupervised learning approach that aims to divide a set of data points into multiple groups. It is a crucial yet demanding subject in machine learning and data mining. Its successful applications span various fields. However, conventional clustering techniques necessitate the consideration of balance significance in specific applications. Therefore, this paper addresses the challenge of imbalanced clustering problems and presents a new method for balanced clustering by utilizing entropy-aware similarity, which can be defined as the degree of balances. We have coined the term, entropy-aware similarity for balanced clustering (EASB), which maximizes balance during clustering by complementary clustering of unbalanced data and incorporating entropy in a novel similarity formula that accounts for both angular differences and distances. The effectiveness of the proposed approach is evaluated on actual melanoma medial data, specifically the International Skin Imaging Collaboration (ISIC) 2019 and 2020 challenge datasets, to demonstrate how it can successfully cluster the data while preserving balance. Lastly, we can confirm that the proposed method exhibited outstanding performance in detecting melanoma, comparing to classical methods.
A Patient-Centric Dataset of Images and Metadata for Identifying Melanomas Using Clinical Context
Aug 07, 2020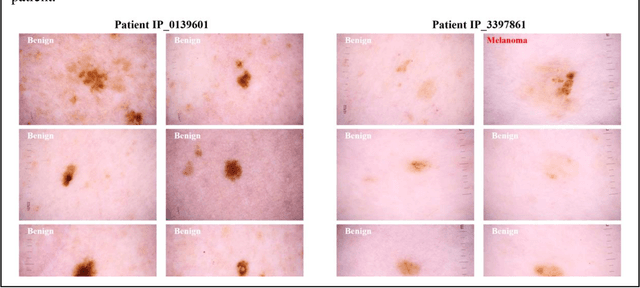
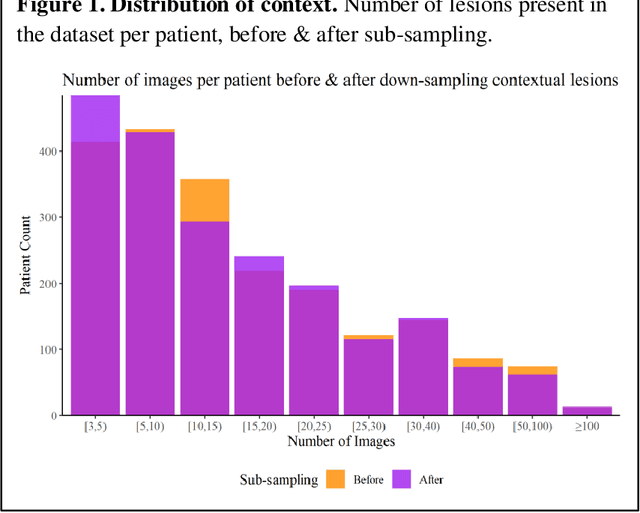
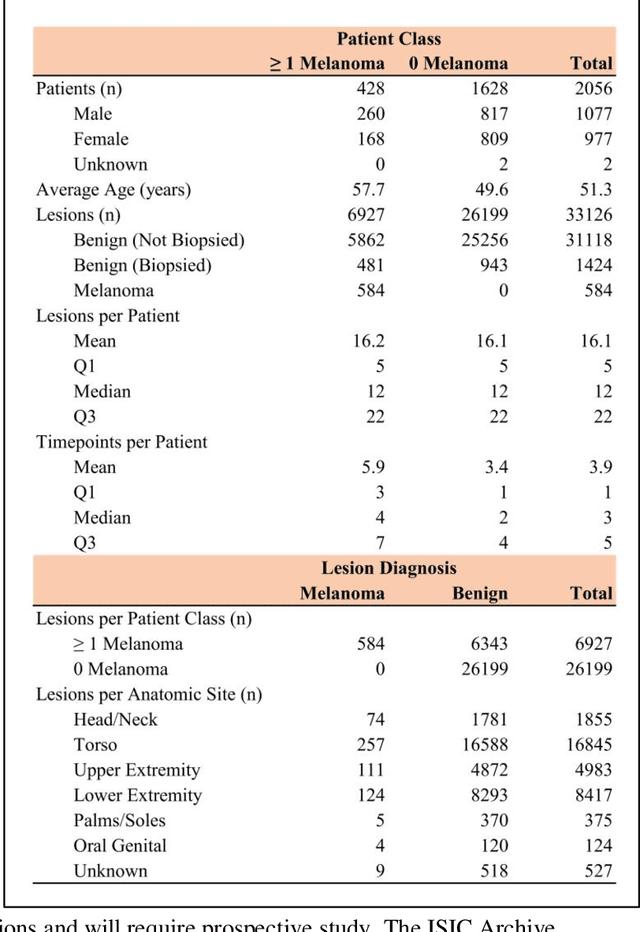
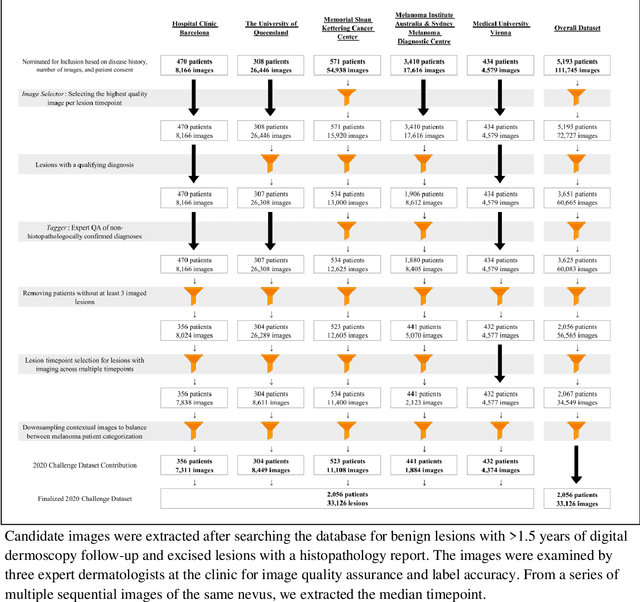
Prior skin image datasets have not addressed patient-level information obtained from multiple skin lesions from the same patient. Though artificial intelligence classification algorithms have achieved expert-level performance in controlled studies examining single images, in practice dermatologists base their judgment holistically from multiple lesions on the same patient. The 2020 SIIM-ISIC Melanoma Classification challenge dataset described herein was constructed to address this discrepancy between prior challenges and clinical practice, providing for each image in the dataset an identifier allowing lesions from the same patient to be mapped to one another. This patient-level contextual information is frequently used by clinicians to diagnose melanoma and is especially useful in ruling out false positives in patients with many atypical nevi. The dataset represents 2,056 patients from three continents with an average of 16 lesions per patient, consisting of 33,126 dermoscopic images and 584 histopathologically confirmed melanomas compared with benign melanoma mimickers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge