Nicholas Kurtansky
On the Role of Calibration in Benchmarking Algorithmic Fairness for Skin Cancer Detection
Nov 10, 2025Abstract:Artificial Intelligence (AI) models have demonstrated expert-level performance in melanoma detection, yet their clinical adoption is hindered by performance disparities across demographic subgroups such as gender, race, and age. Previous efforts to benchmark the performance of AI models have primarily focused on assessing model performance using group fairness metrics that rely on the Area Under the Receiver Operating Characteristic curve (AUROC), which does not provide insights into a model's ability to provide accurate estimates. In line with clinical assessments, this paper addresses this gap by incorporating calibration as a complementary benchmarking metric to AUROC-based fairness metrics. Calibration evaluates the alignment between predicted probabilities and observed event rates, offering deeper insights into subgroup biases. We assess the performance of the leading skin cancer detection algorithm of the ISIC 2020 Challenge on the ISIC 2020 Challenge dataset and the PROVE-AI dataset, and compare it with the second and third place models, focusing on subgroups defined by sex, race (Fitzpatrick Skin Tone), and age. Our findings reveal that while existing models enhance discriminative accuracy, they often over-diagnose risk and exhibit calibration issues when applied to new datasets. This study underscores the necessity for comprehensive model auditing strategies and extensive metadata collection to achieve equitable AI-driven healthcare solutions. All code is publicly available at https://github.com/bdominique/testing_strong_calibration.
* 19 pages, 4 figures. Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2025:027
A Patient-Centric Dataset of Images and Metadata for Identifying Melanomas Using Clinical Context
Aug 07, 2020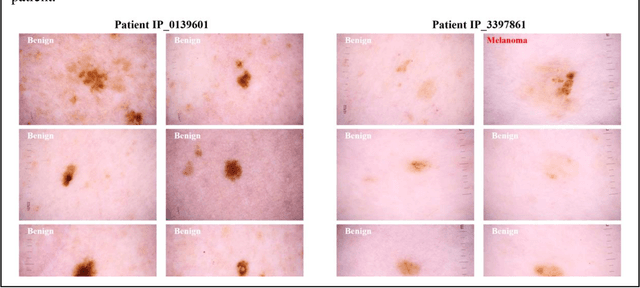
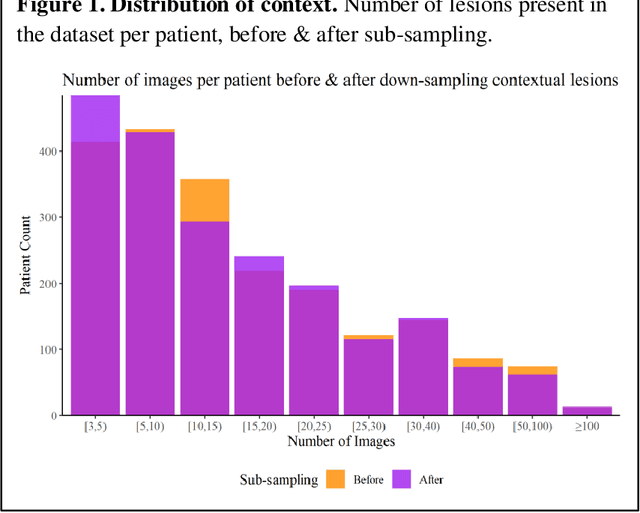
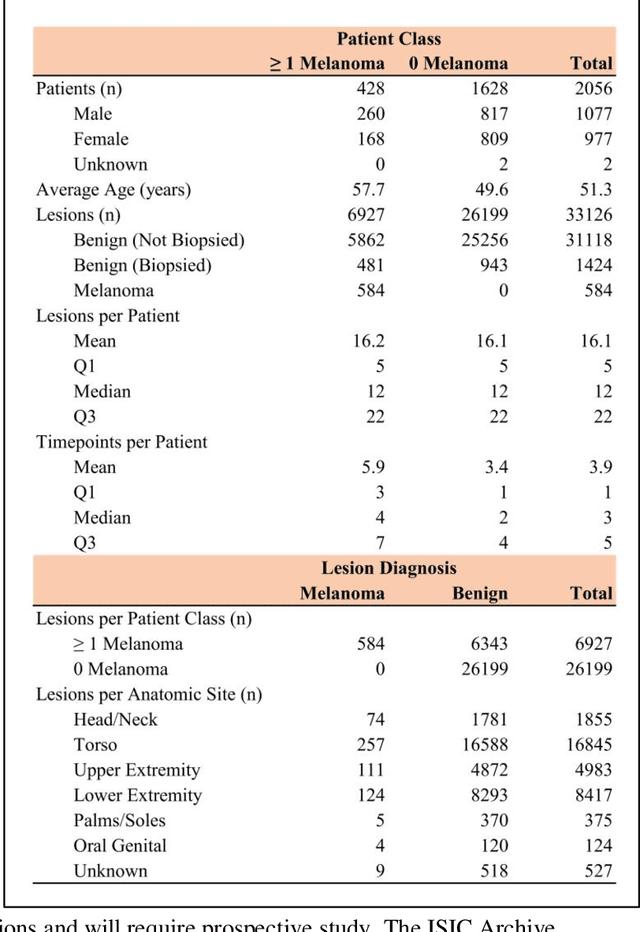
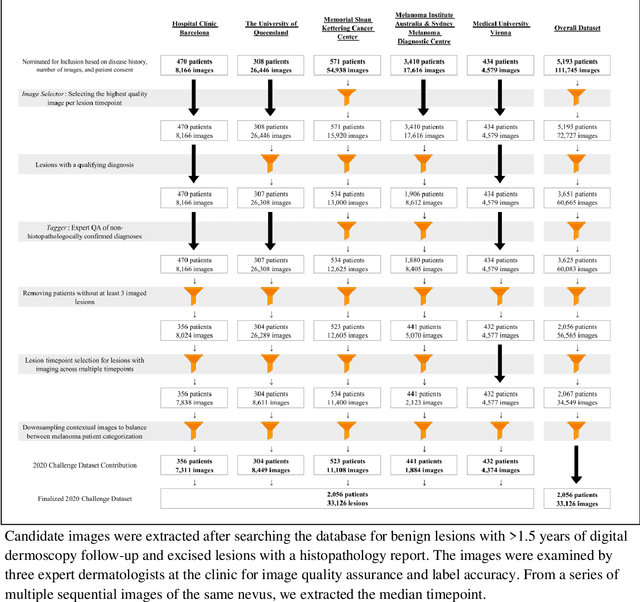
Abstract:Prior skin image datasets have not addressed patient-level information obtained from multiple skin lesions from the same patient. Though artificial intelligence classification algorithms have achieved expert-level performance in controlled studies examining single images, in practice dermatologists base their judgment holistically from multiple lesions on the same patient. The 2020 SIIM-ISIC Melanoma Classification challenge dataset described herein was constructed to address this discrepancy between prior challenges and clinical practice, providing for each image in the dataset an identifier allowing lesions from the same patient to be mapped to one another. This patient-level contextual information is frequently used by clinicians to diagnose melanoma and is especially useful in ruling out false positives in patients with many atypical nevi. The dataset represents 2,056 patients from three continents with an average of 16 lesions per patient, consisting of 33,126 dermoscopic images and 584 histopathologically confirmed melanomas compared with benign melanoma mimickers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge