Zhanghexuan Ji
An End-to-End learnable Flow Regularized Model for Brain Tumor Segmentation
Sep 01, 2021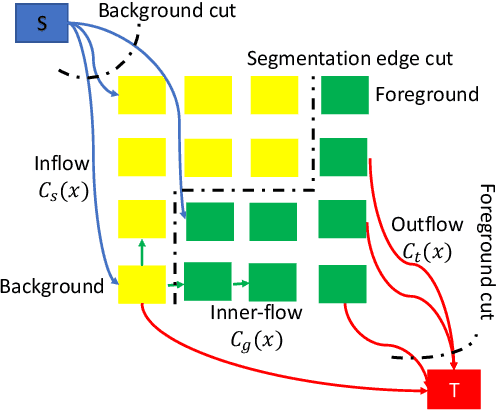
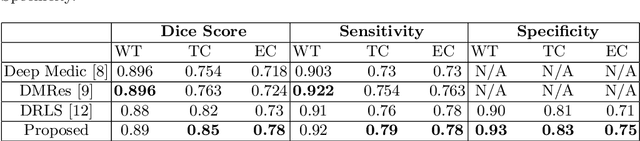
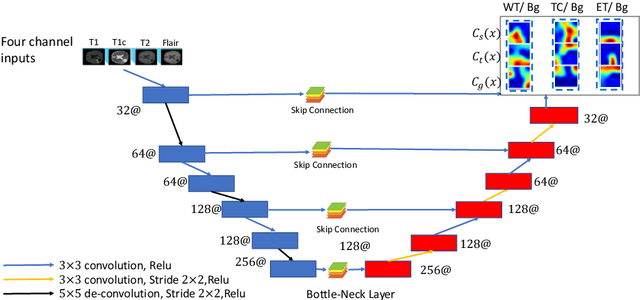
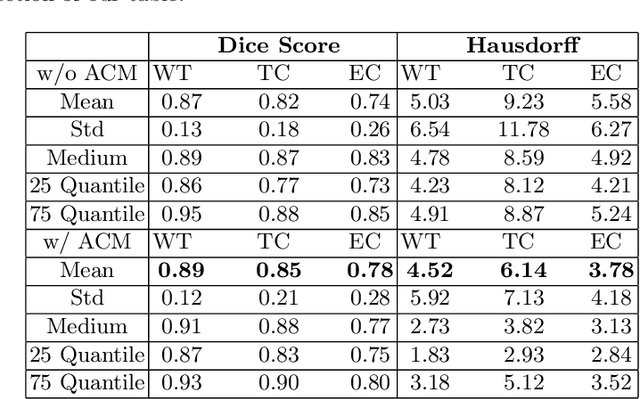
Abstract:Many segmentation tasks for biomedical images can be modeled as the minimization of an energy function and solved by a class of max-flow and min-cut optimization algorithms. However, the segmentation accuracy is sensitive to the contrasting of semantic features of different segmenting objects, as the traditional energy function usually uses hand-crafted features in their energy functions. To address these limitations, we propose to incorporate end-to-end trainable neural network features into the energy functions. Our deep neural network features are extracted from the down-sampling and up-sampling layers with skip-connections of a U-net. In the inference stage, the learned features are fed into the energy functions. And the segmentations are solved in a primal-dual form by ADMM solvers. In the training stage, we train our neural networks by optimizing the energy function in the primal form with regularizations on the min-cut and flow-conservation functions, which are derived from the optimal conditions in the dual form. We evaluate our methods, both qualitatively and quantitatively, in a brain tumor segmentation task. As the energy minimization model achieves a balance on sensitivity and smooth boundaries, we would show how our segmentation contours evolve actively through iterations as ensemble references for doctor diagnosis.
User-Guided Domain Adaptation for Rapid Annotation from User Interactions: A Study on Pathological Liver Segmentation
Sep 05, 2020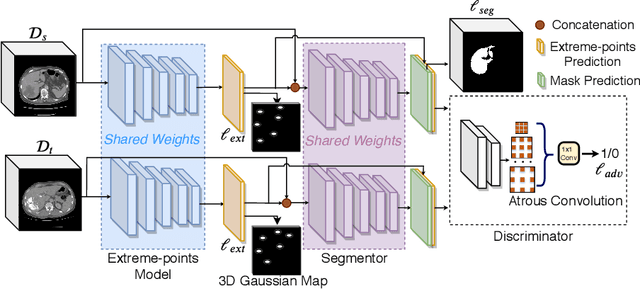
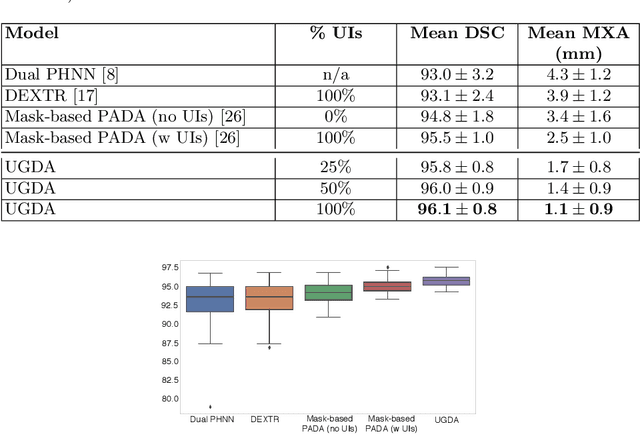

Abstract:Mask-based annotation of medical images, especially for 3D data, is a bottleneck in developing reliable machine learning models. Using minimal-labor user interactions (UIs) to guide the annotation is promising, but challenges remain on best harmonizing the mask prediction with the UIs. To address this, we propose the user-guided domain adaptation (UGDA) framework, which uses prediction-based adversarial domain adaptation (PADA) to model the combined distribution of UIs and mask predictions. The UIs are then used as anchors to guide and align the mask prediction. Importantly, UGDA can both learn from unlabelled data and also model the high-level semantic meaning behind different UIs. We test UGDA on annotating pathological livers using a clinically comprehensive dataset of 927 patient studies. Using only extreme-point UIs, we achieve a mean (worst-case) performance of 96.1%(94.9%), compared to 93.0% (87.0%) for deep extreme points (DEXTR). Furthermore, we also show UGDA can retain this state-of-the-art performance even when only seeing a fraction of available UIs, demonstrating an ability for robust and reliable UI-guided segmentation with extremely minimal labor demands.
Scribble-based Hierarchical Weakly Supervised Learning for Brain Tumor Segmentation
Nov 05, 2019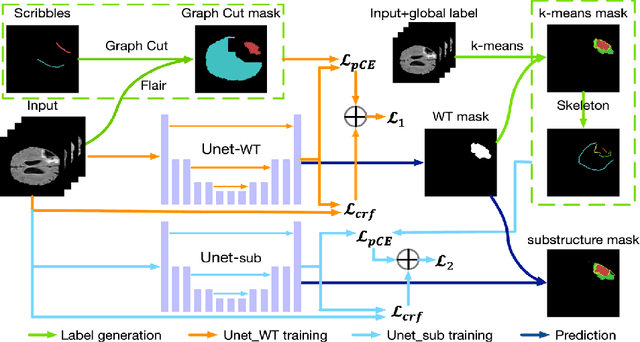
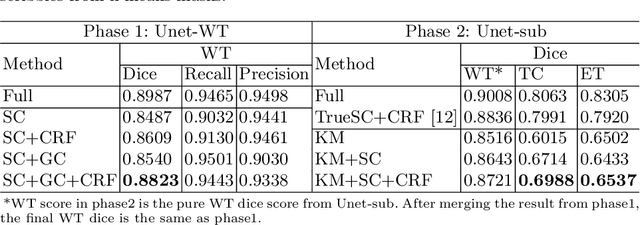
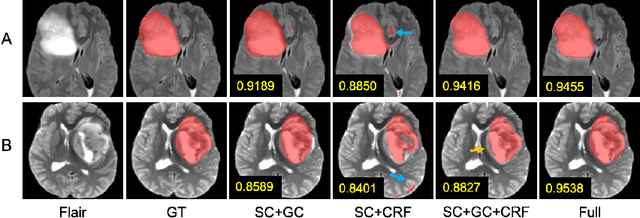
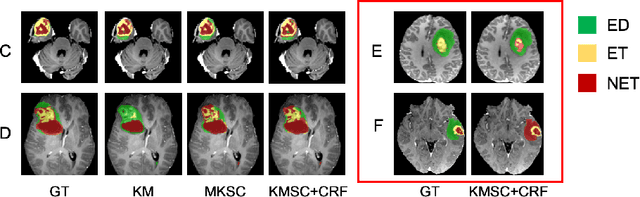
Abstract:The recent state-of-the-art deep learning methods have significantly improved brain tumor segmentation. However, fully supervised training requires a large amount of manually labeled masks, which is highly time-consuming and needs domain expertise. Weakly supervised learning with scribbles provides a good trade-off between model accuracy and the effort of manual labeling. However, for segmenting the hierarchical brain tumor structures, manually labeling scribbles for each substructure could still be demanding. In this paper, we use only two kinds of weak labels, i.e., scribbles on whole tumor and healthy brain tissue, and global labels for the presence of each substructure, to train a deep learning model to segment all the sub-regions. Specifically, we train two networks in two phases: first, we only use whole tumor scribbles to train a whole tumor (WT) segmentation network, which roughly recovers the WT mask of training data; then we cluster the WT region with the guide of global labels. The rough substructure segmentation from clustering is used as weak labels to train the second network. The dense CRF loss is used to refine the weakly supervised segmentation. We evaluate our approach on the BraTS2017 dataset and achieve competitive WT dice score as well as comparable scores on substructure segmentation compared to an upper bound when trained with fully annotated masks.
Neural Style Transfer Improves 3D Cardiovascular MR Image Segmentation on Inconsistent Data
Sep 20, 2019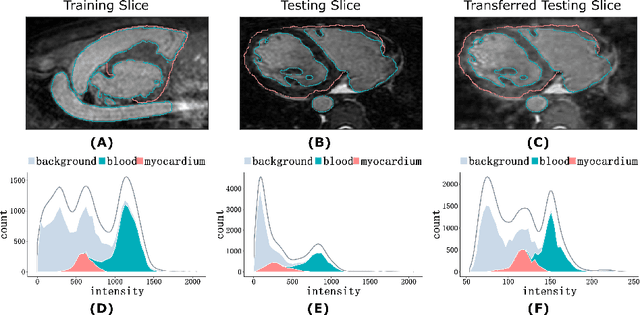
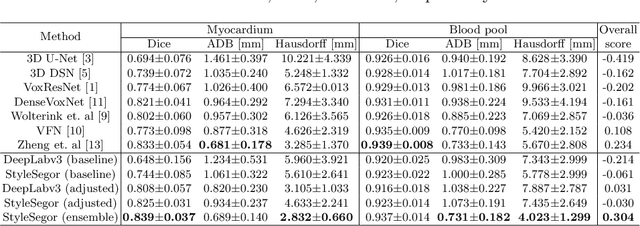
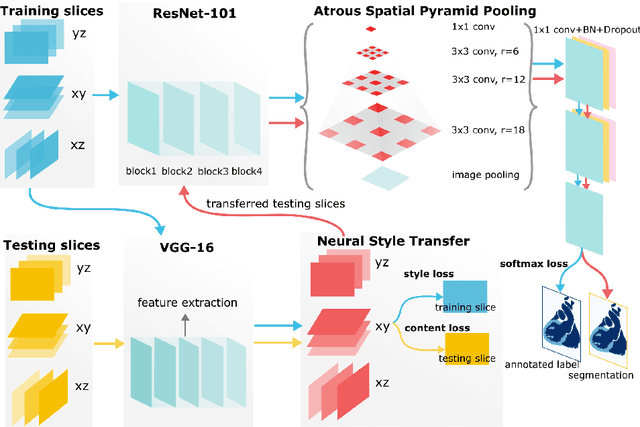
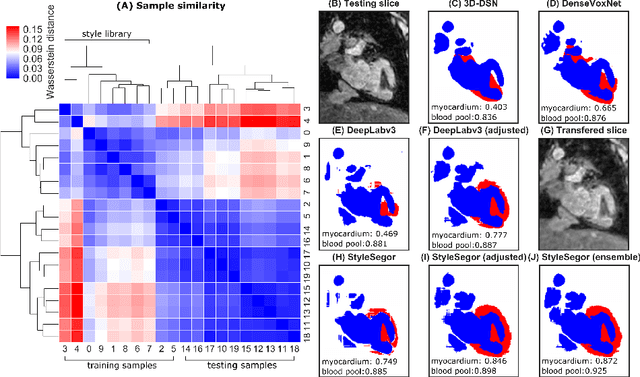
Abstract:Three-dimensional medical image segmentation is one of the most important problems in medical image analysis and plays a key role in downstream diagnosis and treatment. Recent years, deep neural networks have made groundbreaking success in medical image segmentation problem. However, due to the high variance in instrumental parameters, experimental protocols, and subject appearances, the generalization of deep learning models is often hindered by the inconsistency in medical images generated by different machines and hospitals. In this work, we present StyleSegor, an efficient and easy-to-use strategy to alleviate this inconsistency issue. Specifically, neural style transfer algorithm is applied to unlabeled data in order to minimize the differences in image properties including brightness, contrast, texture, etc. between the labeled and unlabeled data. We also apply probabilistic adjustment on the network output and integrate multiple predictions through ensemble learning. On a publicly available whole heart segmentation benchmarking dataset from MICCAI HVSMR 2016 challenge, we have demonstrated an elevated dice accuracy surpassing current state-of-the-art method and notably, an improvement of the total score by 29.91\%. StyleSegor is thus corroborated to be an accurate tool for 3D whole heart segmentation especially on highly inconsistent data, and is available at https://github.com/horsepurve/StyleSegor.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge