Yuyuan Liu
Knowledge Distillation to Ensemble Global and Interpretable Prototype-Based Mammogram Classification Models
Sep 26, 2022Abstract:State-of-the-art (SOTA) deep learning mammogram classifiers, trained with weakly-labelled images, often rely on global models that produce predictions with limited interpretability, which is a key barrier to their successful translation into clinical practice. On the other hand, prototype-based models improve interpretability by associating predictions with training image prototypes, but they are less accurate than global models and their prototypes tend to have poor diversity. We address these two issues with the proposal of BRAIxProtoPNet++, which adds interpretability to a global model by ensembling it with a prototype-based model. BRAIxProtoPNet++ distills the knowledge of the global model when training the prototype-based model with the goal of increasing the classification accuracy of the ensemble. Moreover, we propose an approach to increase prototype diversity by guaranteeing that all prototypes are associated with different training images. Experiments on weakly-labelled private and public datasets show that BRAIxProtoPNet++ has higher classification accuracy than SOTA global and prototype-based models. Using lesion localisation to assess model interpretability, we show BRAIxProtoPNet++ is more effective than other prototype-based models and post-hoc explanation of global models. Finally, we show that the diversity of the prototypes learned by BRAIxProtoPNet++ is superior to SOTA prototype-based approaches.
Translation Consistent Semi-supervised Segmentation for 3D Medical Images
Mar 28, 2022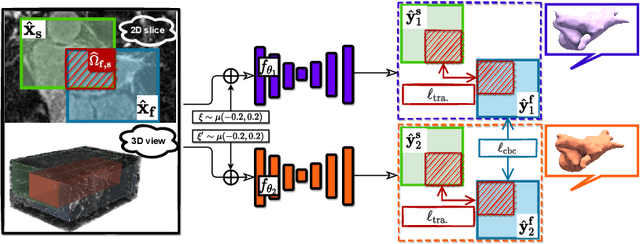
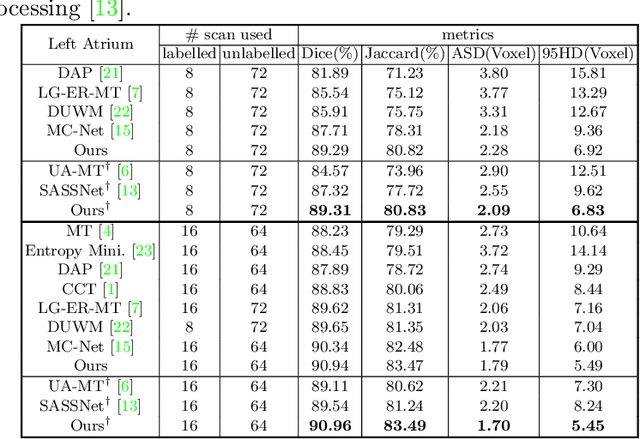
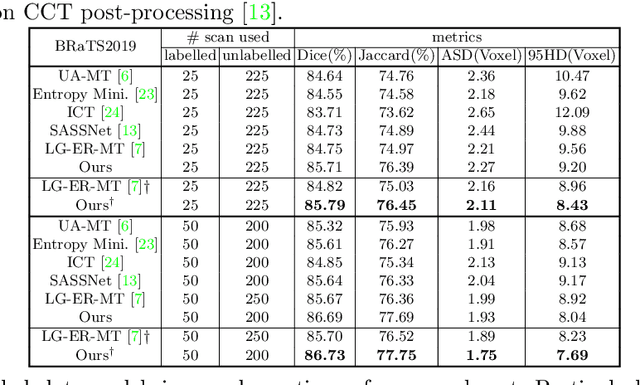
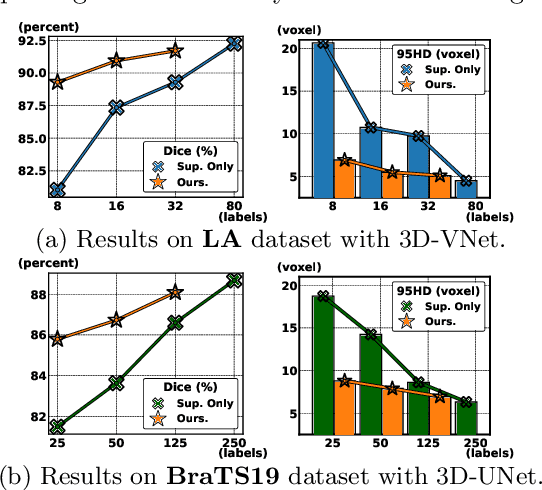
Abstract:3D medical image segmentation methods have been successful, but their dependence on large amounts of voxel-level annotated data is a disadvantage that needs to be addressed given the high cost to obtain such annotation. Semi-supervised learning (SSL) solve this issue by training models with a large unlabelled and a small labelled dataset. The most successful SSL approaches are based on consistency learning that minimises the distance between model responses obtained from perturbed views of the unlabelled data. These perturbations usually keep the spatial input context between views fairly consistent, which may cause the model to learn segmentation patterns from the spatial input contexts instead of the segmented objects. In this paper, we introduce the Translation Consistent Co-training (TraCoCo) which is a consistency learning SSL method that perturbs the input data views by varying their spatial input context, allowing the model to learn segmentation patterns from visual objects. Furthermore, we propose the replacement of the commonly used mean squared error (MSE) semi-supervised loss by a new Cross-model confident Binary Cross entropy (CBC) loss, which improves training convergence and keeps the robustness to co-training pseudo-labelling mistakes. We also extend CutMix augmentation to 3D SSL to further improve generalisation. Our TraCoCo shows state-of-the-art results for the Left Atrium (LA) and Brain Tumor Segmentation (BRaTS19) datasets with different backbones. Our code is available at https://github.com/yyliu01/TraCoCo.
Contrastive Transformer-based Multiple Instance Learning for Weakly Supervised Polyp Frame Detection
Mar 23, 2022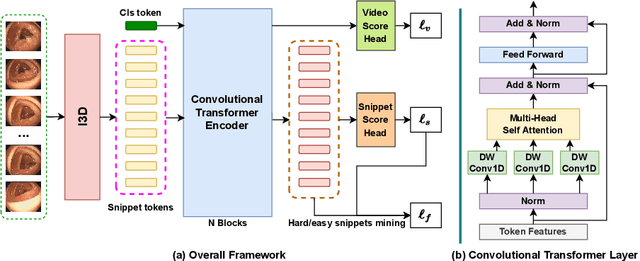
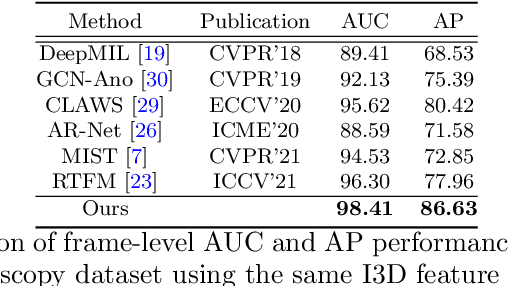
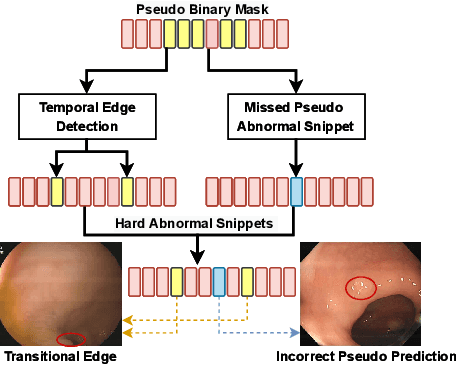

Abstract:Current polyp detection methods from colonoscopy videos use exclusively normal (i.e., healthy) training images, which i) ignore the importance of temporal information in consecutive video frames, and ii) lack knowledge about the polyps. Consequently, they often have high detection errors, especially on challenging polyp cases (e.g., small, flat, or partially visible polyps). In this work, we formulate polyp detection as a weakly-supervised anomaly detection task that uses video-level labelled training data to detect frame-level polyps. In particular, we propose a novel convolutional transformer-based multiple instance learning method designed to identify abnormal frames (i.e., frames with polyps) from anomalous videos (i.e., videos containing at least one frame with polyp). In our method, local and global temporal dependencies are seamlessly captured while we simultaneously optimise video and snippet-level anomaly scores. A contrastive snippet mining method is also proposed to enable an effective modelling of the challenging polyp cases. The resulting method achieves a detection accuracy that is substantially better than current state-of-the-art approaches on a new large-scale colonoscopy video dataset introduced in this work.
Unsupervised Anomaly Detection in Medical Images with a Memory-augmented Multi-level Cross-attentional Masked Autoencoder
Mar 22, 2022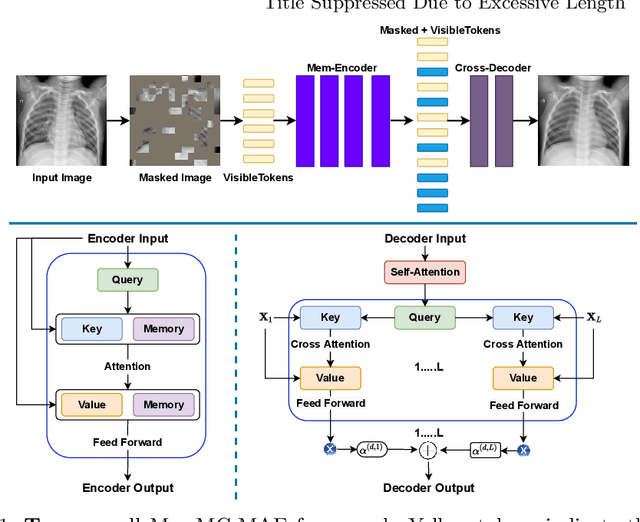



Abstract:Unsupervised anomaly detection (UAD) aims to find anomalous images by optimising a detector using a training set that contains only normal images. UAD approaches can be based on reconstruction methods, self-supervised approaches, and Imagenet pre-trained models. Reconstruction methods, which detect anomalies from image reconstruction errors, are advantageous because they do not rely on the design of problem-specific pretext tasks needed by self-supervised approaches, and on the unreliable translation of models pre-trained from non-medical datasets. However, reconstruction methods may fail because they can have low reconstruction errors even for anomalous images. In this paper, we introduce a new reconstruction-based UAD approach that addresses this low-reconstruction error issue for anomalous images. Our UAD approach, the memory-augmented multi-level cross-attentional masked autoencoder (MemMC-MAE), is a transformer-based approach, consisting of a novel memory-augmented self-attention operator for the encoder and a new multi-level cross-attention operator for the decoder. MemMC-MAE masks large parts of the input image during its reconstruction, reducing the risk that it will produce low reconstruction errors because anomalies are likely to be masked and cannot be reconstructed. However, when the anomaly is not masked, then the normal patterns stored in the encoder's memory combined with the decoder's multi-level cross-attention will constrain the accurate reconstruction of the anomaly. We show that our method achieves SOTA anomaly detection and localisation on colonoscopy and Covid-19 Chest X-ray datasets.
Semantic-guided Image Virtual Attribute Learning for Noisy Multi-label Chest X-ray Classification
Mar 03, 2022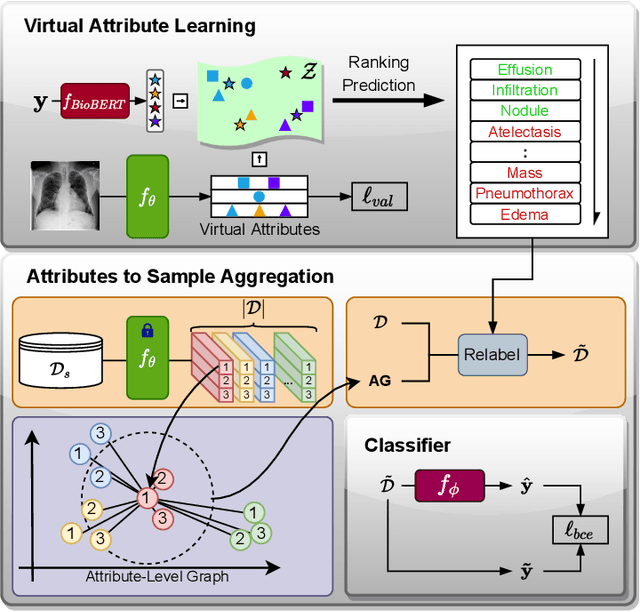
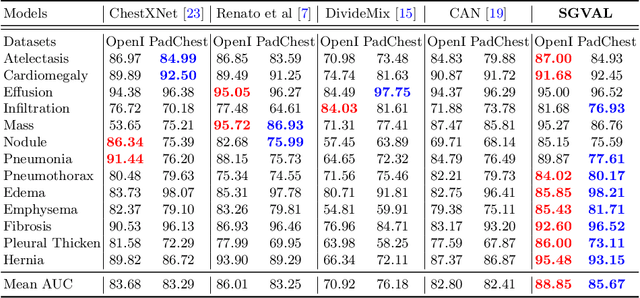
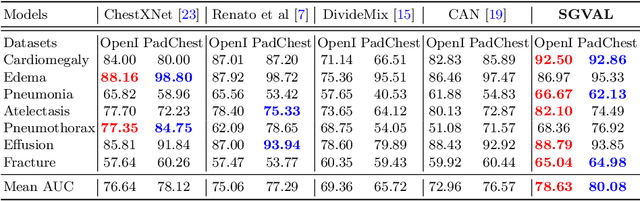
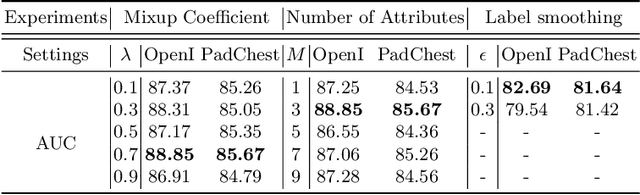
Abstract:Deep learning methods have shown outstanding classification accuracy in medical image analysis problems, which is largely attributed to the availability of large datasets manually annotated with clean labels. However, such manual annotation can be expensive to obtain for large datasets, so we may rely on machine-generated noisy labels. Many Chest X-ray (CXR) classifiers are modelled from datasets with machine-generated labels, but their training procedure is in general not robust to the presence of noisy-label samples and can overfit those samples to produce sub-optimal solutions. Furthermore, CXR datasets are mostly multi-label, so current noisy-label learning methods designed for multi-class problems cannot be easily adapted. To address such noisy multi-label CXR learning problem, we propose a new learning method based on estimating image virtual attributes using semantic information from the label to assist in the identification and correction of noisy multi-labels from training samples. Our experiments on diverse noisy multi-label training sets and clean testing sets show that our model has state-of-the-art accuracy and robustness across all datasets.
ACPL: Anti-curriculum Pseudo-labelling for Semi-supervised Medical Image Classification
Dec 19, 2021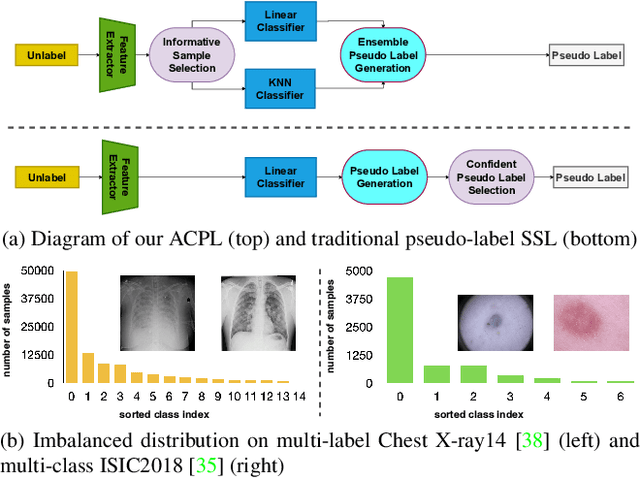
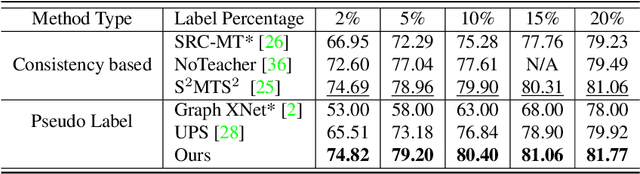
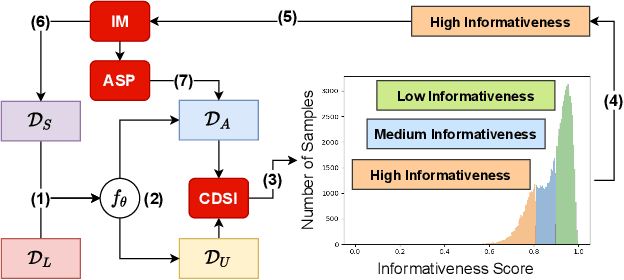
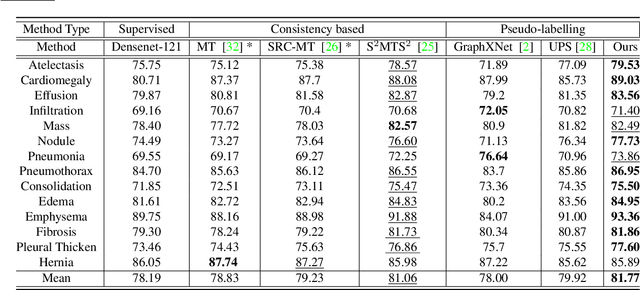
Abstract:Effective semi-supervised learning (SSL) in medical im-age analysis (MIA) must address two challenges: 1) workeffectively on both multi-class (e.g., lesion classification)and multi-label (e.g., multiple-disease diagnosis) problems,and 2) handle imbalanced learning (because of the highvariance in disease prevalence). One strategy to explorein SSL MIA is based on the pseudo labelling strategy, butit has a few shortcomings. Pseudo-labelling has in generallower accuracy than consistency learning, it is not specifi-cally design for both multi-class and multi-label problems,and it can be challenged by imbalanced learning. In this paper, unlike traditional methods that select confident pseudo label by threshold, we propose a new SSL algorithm, called anti-curriculum pseudo-labelling (ACPL), which introduces novel techniques to select informative unlabelled samples, improving training balance and allowing the model to work for both multi-label and multi-class problems, and to estimate pseudo labels by an accurate ensemble of classifiers(improving pseudo label accuracy). We run extensive experiments to evaluate ACPL on two public medical image classification benchmarks: Chest X-Ray14 for thorax disease multi-label classification and ISIC2018 for skin lesion multi-class classification. Our method outperforms previous SOTA SSL methods on both datasets.
Perturbed and Strict Mean Teachers for Semi-supervised Semantic Segmentation
Nov 25, 2021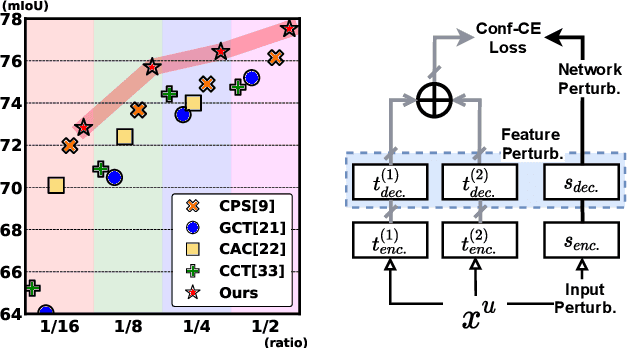
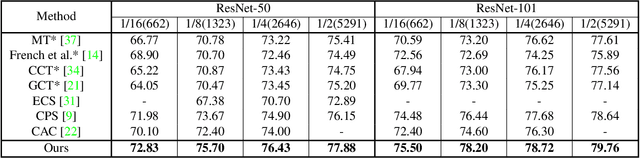
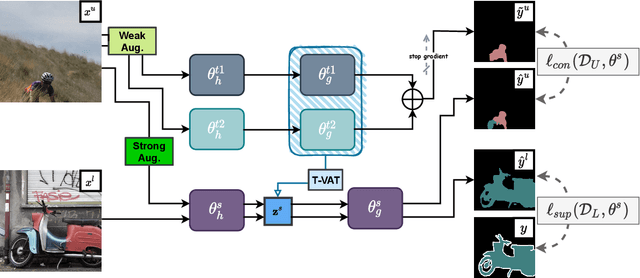
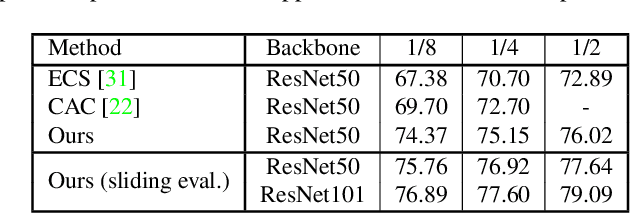
Abstract:Consistency learning using input image, feature, or network perturbations has shown remarkable results in semi-supervised semantic segmentation, but this approach can be seriously affected by inaccurate predictions of unlabelled training images. There are two consequences of these inaccurate predictions: 1) the training based on the ``strict'' cross-entropy (CE) loss can easily overfit prediction mistakes, leading to confirmation bias; and 2) the perturbations applied to these inaccurate predictions will use potentially erroneous predictions as training signals, degrading consistency learning. In this paper, we address the prediction accuracy problem of consistency learning methods with novel extensions of the mean-teacher (MT) model, which include a new auxiliary teacher, and the replacement of MT's mean square error (MSE) by a stricter confidence-weighted cross-entropy (Conf-CE) loss. The accurate prediction by this model allows us to use a challenging combination of network, input data and feature perturbations to improve the consistency learning generalisation, where the feature perturbations consist of a new adversarial perturbation. Results on public benchmarks show that our approach achieves remarkable improvements over the previous SOTA methods in the field.
Pixel-wise Energy-biased Abstention Learning for Anomaly Segmentation on Complex Urban Driving Scenes
Nov 24, 2021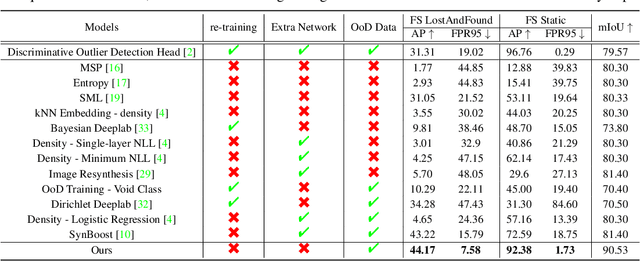
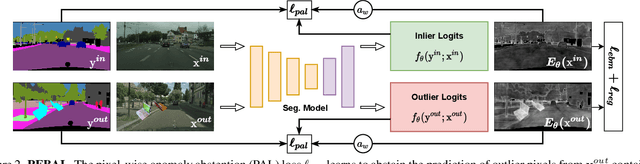
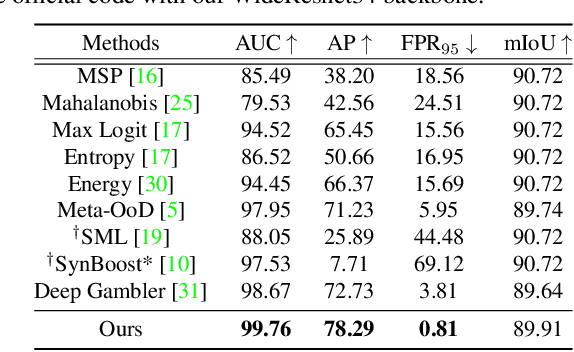
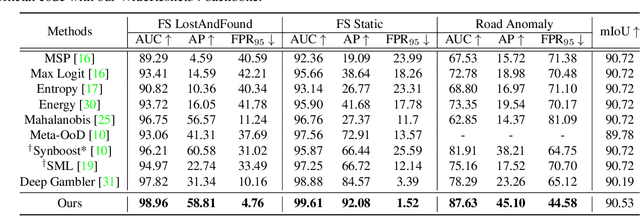
Abstract:State-of-the-art (SOTA) anomaly segmentation approaches on complex urban driving scenes explore pixel-wise classification uncertainty learned from outlier exposure, or external reconstruction models. However, previous uncertainty approaches that directly associate high uncertainty to anomaly may sometimes lead to incorrect anomaly predictions, and external reconstruction models tend to be too inefficient for real-time self-driving embedded systems. In this paper, we propose a new anomaly segmentation method, named pixel-wise energy-biased abstention learning (PEBAL), that explores pixel-wise abstention learning (AL) with a model that learns an adaptive pixel-level anomaly class, and an energy-based model (EBM) that learns inlier pixel distribution. More specifically, PEBAL is based on a non-trivial joint training of EBM and AL, where EBM is trained to output high-energy for anomaly pixels (from outlier exposure) and AL is trained such that these high-energy pixels receive adaptive low penalty for being included to the anomaly class. We extensively evaluate PEBAL against the SOTA and show that it achieves the best performance across four benchmarks. Code is available at https://github.com/tianyu0207/PEBAL.
Multi-centred Strong Augmentation via Contrastive Learning for Unsupervised Lesion Detection and Segmentation
Sep 03, 2021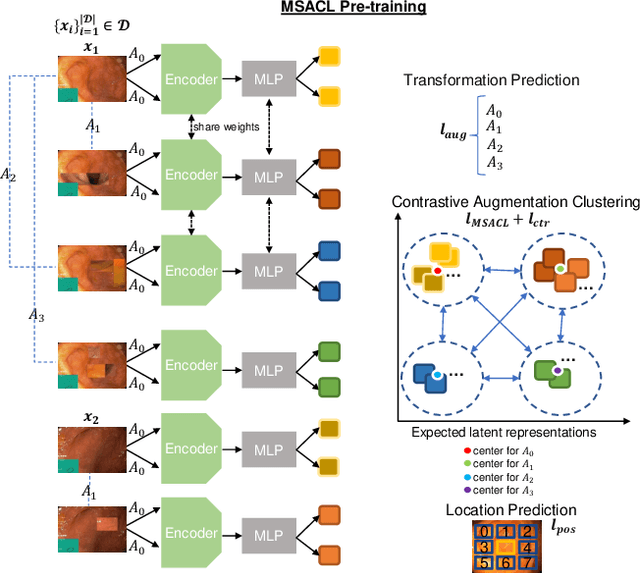
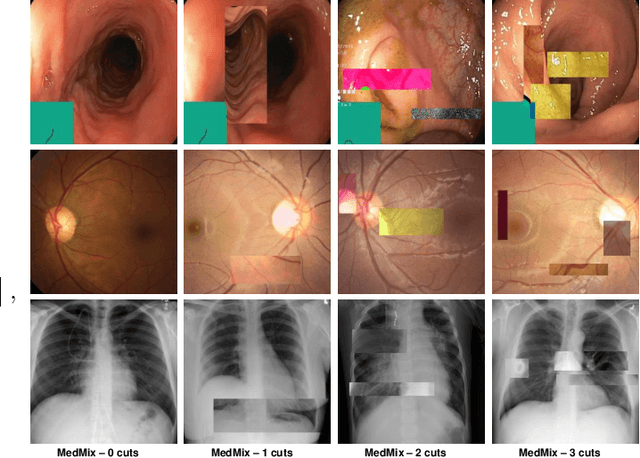
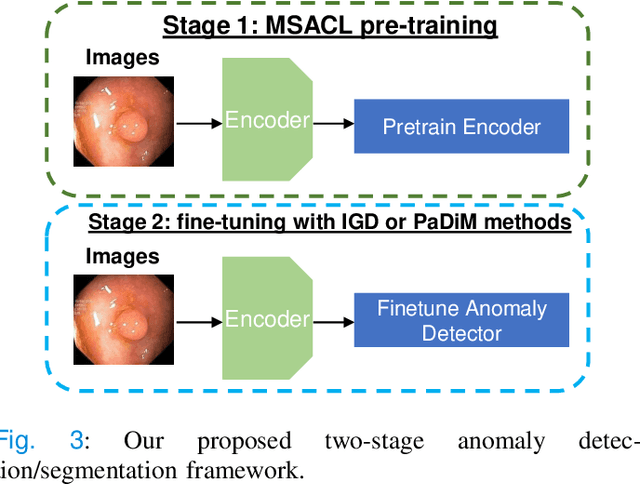
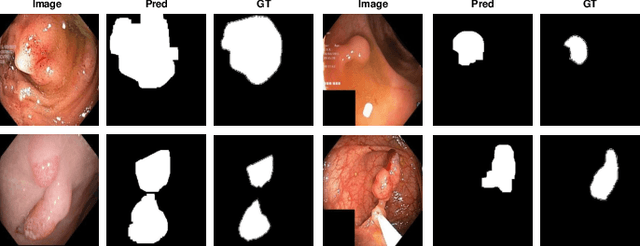
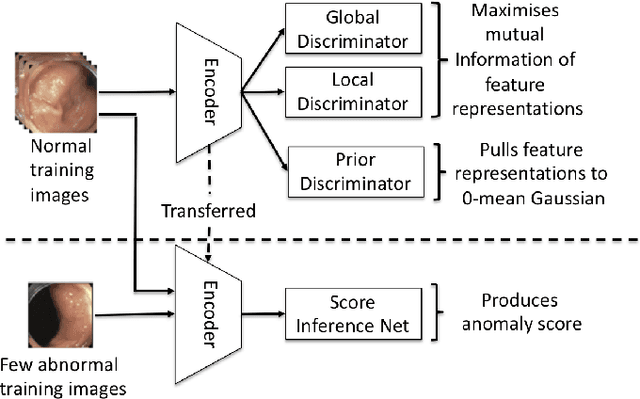
Abstract:The scarcity of high quality medical image annotations hinders the implementation of accurate clinical applications for detecting and segmenting abnormal lesions. To mitigate this issue, the scientific community is working on the development of unsupervised anomaly detection (UAD) systems that learn from a training set containing only normal (i.e., healthy) images, where abnormal samples (i.e., unhealthy) are detected and segmented based on how much they deviate from the learned distribution of normal samples. One significant challenge faced by UAD methods is how to learn effective low-dimensional image representations that are sensitive enough to detect and segment abnormal lesions of varying size, appearance and shape. To address this challenge, we propose a novel self-supervised UAD pre-training algorithm, named Multi-centred Strong Augmentation via Contrastive Learning (MSACL). MSACL learns representations by separating several types of strong and weak augmentations of normal image samples, where the weak augmentations represent normal images and strong augmentations denote synthetic abnormal images. To produce such strong augmentations, we introduce MedMix, a novel data augmentation strategy that creates new training images with realistic looking lesions (i.e., anomalies) in normal images. The pre-trained representations from MSACL are generic and can be used to improve the efficacy of different types of off-the-shelf state-of-the-art (SOTA) UAD models. Comprehensive experimental results show that the use of MSACL largely improves these SOTA UAD models on four medical imaging datasets from diverse organs, namely colonoscopy, fundus screening and covid-19 chest-ray datasets.
Detecting, Localising and Classifying Polyps from Colonoscopy Videos using Deep Learning
Jan 09, 2021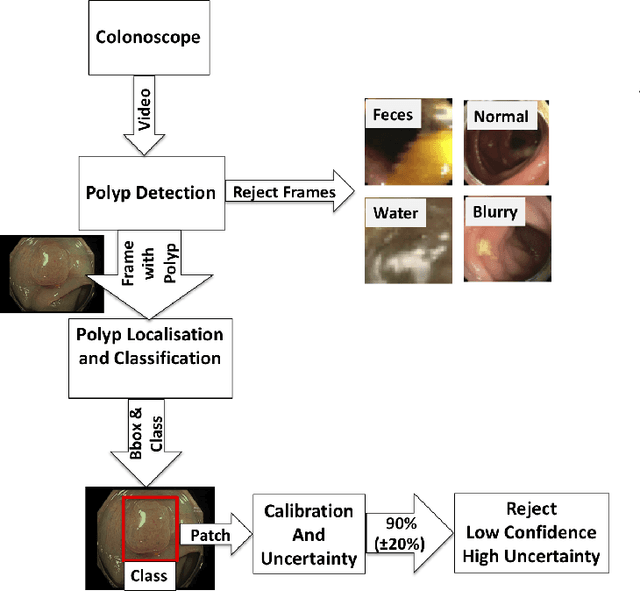
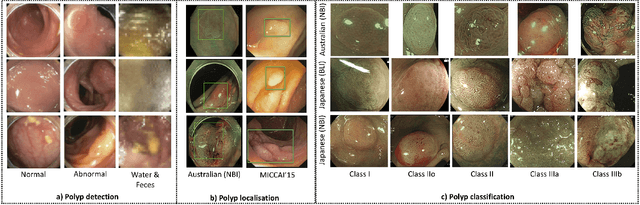
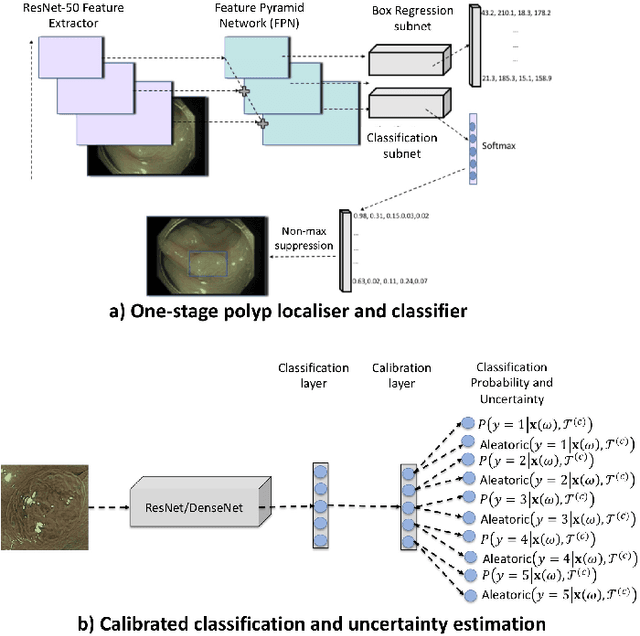
Abstract:In this paper, we propose and analyse a system that can automatically detect, localise and classify polyps from colonoscopy videos. The detection of frames with polyps is formulated as a few-shot anomaly classification problem, where the training set is highly imbalanced with the large majority of frames consisting of normal images and a small minority comprising frames with polyps. Colonoscopy videos may contain blurry images and frames displaying feces and water jet sprays to clean the colon -- such frames can mistakenly be detected as anomalies, so we have implemented a classifier to reject these two types of frames before polyp detection takes place. Next, given a frame containing a polyp, our method localises (with a bounding box around the polyp) and classifies it into five different classes. Furthermore, we study a method to improve the reliability and interpretability of the classification result using uncertainty estimation and classification calibration. Classification uncertainty and calibration not only help improve classification accuracy by rejecting low-confidence and high-uncertain results, but can be used by doctors to decide how to decide on the classification of a polyp. All the proposed detection, localisation and classification methods are tested using large data sets and compared with relevant baseline approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge