Yunqi Hong
Understanding Reward Hacking in Text-to-Image Reinforcement Learning
Jan 06, 2026Abstract:Reinforcement learning (RL) has become a standard approach for post-training large language models and, more recently, for improving image generation models, which uses reward functions to enhance generation quality and human preference alignment. However, existing reward designs are often imperfect proxies for true human judgment, making models prone to reward hacking--producing unrealistic or low-quality images that nevertheless achieve high reward scores. In this work, we systematically analyze reward hacking behaviors in text-to-image (T2I) RL post-training. We investigate how both aesthetic/human preference rewards and prompt-image consistency rewards individually contribute to reward hacking and further show that ensembling multiple rewards can only partially mitigate this issue. Across diverse reward models, we identify a common failure mode: the generation of artifact-prone images. To address this, we propose a lightweight and adaptive artifact reward model, trained on a small curated dataset of artifact-free and artifact-containing samples. This model can be integrated into existing RL pipelines as an effective regularizer for commonly used reward models. Experiments demonstrate that incorporating our artifact reward significantly improves visual realism and reduces reward hacking across multiple T2I RL setups, demonstrating the effectiveness of lightweight reward augment serving as a safeguard against reward hacking.
Adaptive Diagnostic Reasoning Framework for Pathology with Multimodal Large Language Models
Nov 15, 2025Abstract:AI tools in pathology have improved screening throughput, standardized quantification, and revealed prognostic patterns that inform treatment. However, adoption remains limited because most systems still lack the human-readable reasoning needed to audit decisions and prevent errors. We present RECAP-PATH, an interpretable framework that establishes a self-learning paradigm, shifting off-the-shelf multimodal large language models from passive pattern recognition to evidence-linked diagnostic reasoning. At its core is a two-phase learning process that autonomously derives diagnostic criteria: diversification expands pathology-style explanations, while optimization refines them for accuracy. This self-learning approach requires only small labeled sets and no white-box access or weight updates to generate cancer diagnoses. Evaluated on breast and prostate datasets, RECAP-PATH produced rationales aligned with expert assessment and delivered substantial gains in diagnostic accuracy over baselines. By uniting visual understanding with reasoning, RECAP-PATH provides clinically trustworthy AI and demonstrates a generalizable path toward evidence-linked interpretation.
Uncertainty-Guided Selective Adaptation Enables Cross-Platform Predictive Fluorescence Microscopy
Nov 15, 2025Abstract:Deep learning is transforming microscopy, yet models often fail when applied to images from new instruments or acquisition settings. Conventional adversarial domain adaptation (ADDA) retrains entire networks, often disrupting learned semantic representations. Here, we overturn this paradigm by showing that adapting only the earliest convolutional layers, while freezing deeper layers, yields reliable transfer. Building on this principle, we introduce Subnetwork Image Translation ADDA with automatic depth selection (SIT-ADDA-Auto), a self-configuring framework that integrates shallow-layer adversarial alignment with predictive uncertainty to automatically select adaptation depth without target labels. We demonstrate robustness via multi-metric evaluation, blinded expert assessment, and uncertainty-depth ablations. Across exposure and illumination shifts, cross-instrument transfer, and multiple stains, SIT-ADDA improves reconstruction and downstream segmentation over full-encoder adaptation and non-adversarial baselines, with reduced drift of semantic features. Our results provide a design rule for label-free adaptation in microscopy and a recipe for field settings; the code is publicly available.
Graph Neural Diffusion Networks for Semi-supervised Learning
Jan 24, 2022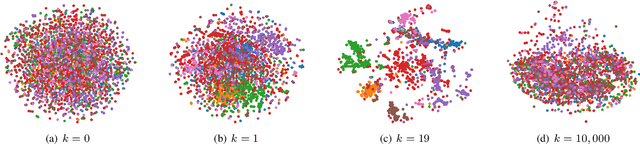
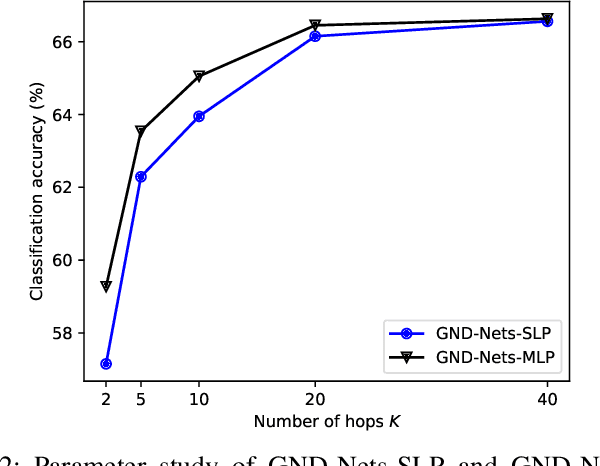
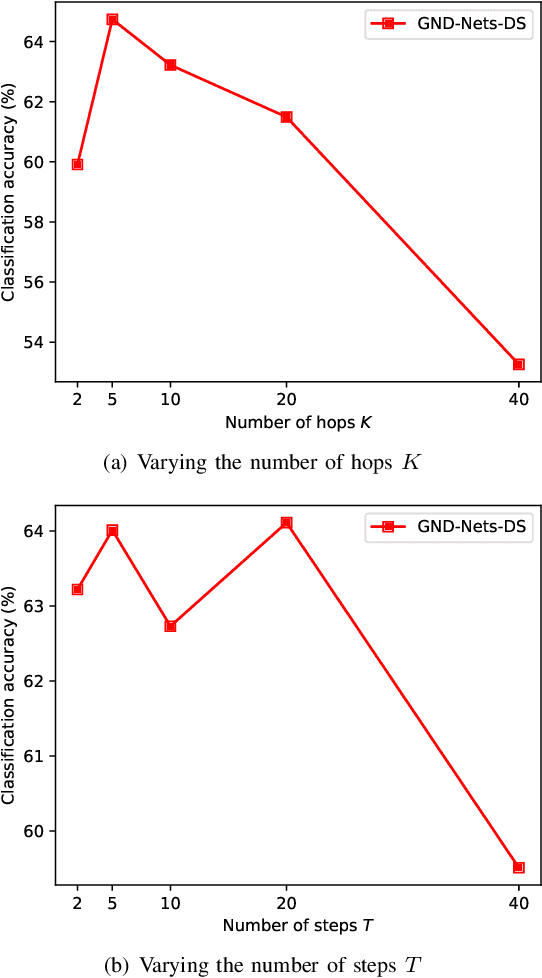
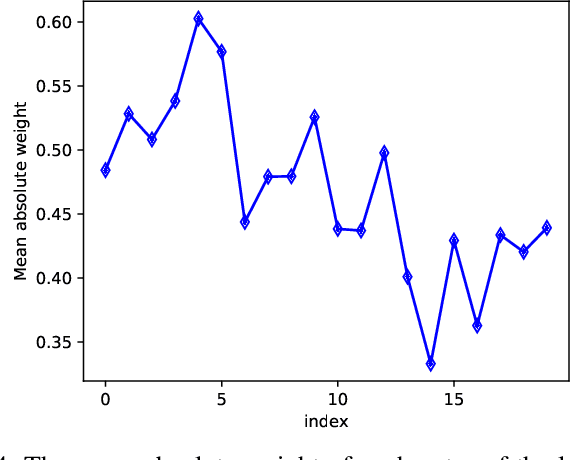
Abstract:Graph Convolutional Networks (GCN) is a pioneering model for graph-based semi-supervised learning. However, GCN does not perform well on sparsely-labeled graphs. Its two-layer version cannot effectively propagate the label information to the whole graph structure (i.e., the under-smoothing problem) while its deep version over-smoothens and is hard to train (i.e., the over-smoothing problem). To solve these two issues, we propose a new graph neural network called GND-Nets (for Graph Neural Diffusion Networks) that exploits the local and global neighborhood information of a vertex in a single layer. Exploiting the shallow network mitigates the over-smoothing problem while exploiting the local and global neighborhood information mitigates the under-smoothing problem. The utilization of the local and global neighborhood information of a vertex is achieved by a new graph diffusion method called neural diffusions, which integrate neural networks into the conventional linear and nonlinear graph diffusions. The adoption of neural networks makes neural diffusions adaptable to different datasets. Extensive experiments on various sparsely-labeled graphs verify the effectiveness and efficiency of GND-Nets compared to state-of-the-art approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge