Yuan Ji
Cross-channel Perception Learning for H&E-to-IHC Virtual Staining
Jun 09, 2025



Abstract:With the rapid development of digital pathology, virtual staining has become a key technology in multimedia medical information systems, offering new possibilities for the analysis and diagnosis of pathological images. However, existing H&E-to-IHC studies often overlook the cross-channel correlations between cell nuclei and cell membranes. To address this issue, we propose a novel Cross-Channel Perception Learning (CCPL) strategy. Specifically, CCPL first decomposes HER2 immunohistochemical staining into Hematoxylin and DAB staining channels, corresponding to cell nuclei and cell membranes, respectively. Using the pathology foundation model Gigapath's Tile Encoder, CCPL extracts dual-channel features from both the generated and real images and measures cross-channel correlations between nuclei and membranes. The features of the generated and real stained images, obtained through the Tile Encoder, are also used to calculate feature distillation loss, enhancing the model's feature extraction capabilities without increasing the inference burden. Additionally, CCPL performs statistical analysis on the focal optical density maps of both single channels to ensure consistency in staining distribution and intensity. Experimental results, based on quantitative metrics such as PSNR, SSIM, PCC, and FID, along with professional evaluations from pathologists, demonstrate that CCPL effectively preserves pathological features, generates high-quality virtual stained images, and provides robust support for automated pathological diagnosis using multimedia medical data.
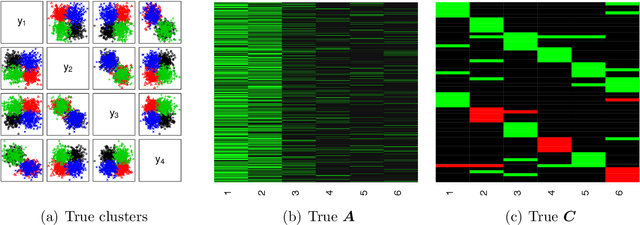
PAM: Plaid Atoms Model for Bayesian Nonparametric Analysis of Grouped Data
Apr 28, 2023Abstract:We consider dependent clustering of observations in groups. The proposed model, called the plaid atoms model (PAM), estimates a set of clusters for each group and allows some clusters to be either shared with other groups or uniquely possessed by the group. PAM is based on an extension to the well-known stick-breaking process by adding zero as a possible value for the cluster weights, resulting in a zero-augmented beta (ZAB) distribution in the model. As a result, ZAB allows some cluster weights to be exactly zero in multiple groups, thereby enabling shared and unique atoms across groups. We explore theoretical properties of PAM and show its connection to known Bayesian nonparametric models. We propose an efficient slice sampler for posterior inference. Minor extensions of the proposed model for multivariate or count data are presented. Simulation studies and applications using real-world datasets illustrate the model's desirable performance.
Consensus Monte Carlo for Random Subsets using Shared Anchors
Jun 28, 2019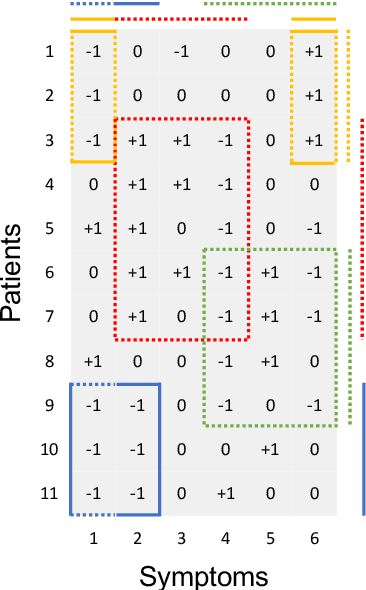

Abstract:We present a consensus Monte Carlo algorithm that scales existing Bayesian nonparametric models for clustering and feature allocation to big data. The algorithm is valid for any prior on random subsets such as partitions and latent feature allocation, under essentially any sampling model. Motivated by three case studies, we focus on clustering induced by a Dirichlet process mixture sampling model, inference under an Indian buffet process prior with a binomial sampling model, and with a categorical sampling model. We assess the proposed algorithm with simulation studies and show results for inference with three datasets: an MNIST image dataset, a dataset of pancreatic cancer mutations, and a large set of electronic health records (EHR). Supplementary materials for this article are available online.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge