Yanting Liu
Protein Design with Dynamic Protein Vocabulary
May 25, 2025



Abstract:Protein design is a fundamental challenge in biotechnology, aiming to design novel sequences with specific functions within the vast space of possible proteins. Recent advances in deep generative models have enabled function-based protein design from textual descriptions, yet struggle with structural plausibility. Inspired by classical protein design methods that leverage natural protein structures, we explore whether incorporating fragments from natural proteins can enhance foldability in generative models. Our empirical results show that even random incorporation of fragments improves foldability. Building on this insight, we introduce ProDVa, a novel protein design approach that integrates a text encoder for functional descriptions, a protein language model for designing proteins, and a fragment encoder to dynamically retrieve protein fragments based on textual functional descriptions. Experimental results demonstrate that our approach effectively designs protein sequences that are both functionally aligned and structurally plausible. Compared to state-of-the-art models, ProDVa achieves comparable function alignment using less than 0.04% of the training data, while designing significantly more well-folded proteins, with the proportion of proteins having pLDDT above 70 increasing by 7.38% and those with PAE below 10 increasing by 9.6%.
Generation with Dynamic Vocabulary
Oct 11, 2024



Abstract:We introduce a new dynamic vocabulary for language models. It can involve arbitrary text spans during generation. These text spans act as basic generation bricks, akin to tokens in the traditional static vocabularies. We show that, the ability to generate multi-tokens atomically improve both generation quality and efficiency (compared to the standard language model, the MAUVE metric is increased by 25%, the latency is decreased by 20%). The dynamic vocabulary can be deployed in a plug-and-play way, thus is attractive for various downstream applications. For example, we demonstrate that dynamic vocabulary can be applied to different domains in a training-free manner. It also helps to generate reliable citations in question answering tasks (substantially enhancing citation results without compromising answer accuracy).
Efficient refinements on YOLOv3 for real-time detection and assessment of diabetic foot Wagner grades
Jun 04, 2020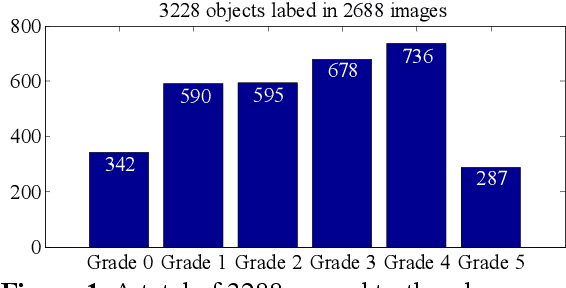
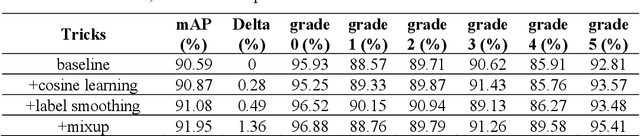
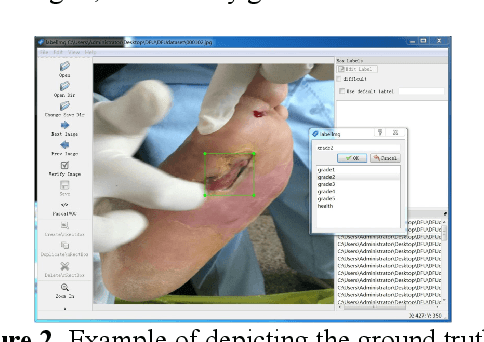
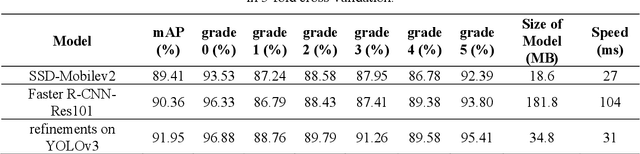
Abstract:Currently, the screening of Wagner grades of diabetic feet (DF) still relies on professional podiatrists. However, in less-developed countries, podiatrists are scarce, which led to the majority of undiagnosed patients. In this study, we proposed the real-time detection and location method for Wagner grades of DF based on refinements on YOLOv3. We collected 2,688 data samples and implemented several methods, such as a visual coherent image mixup, label smoothing, and training scheduler revamping, based on the ablation study. The experimental results suggested that the refinements on YOLOv3 achieved an accuracy of 91.95% and the inference speed of a single picture reaches 31ms with the NVIDIA Tesla V100. To test the performance of the model on a smartphone, we deployed the refinements on YOLOv3 models on an Android 9 system smartphone. This work has the potential to lead to a paradigm shift for clinical treatment of the DF in the future, to provide an effective healthcare solution for DF tissue analysis and healing status.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge