Yanan Zhu
MolSculpt: Sculpting 3D Molecular Geometries from Chemical Syntax
Dec 09, 2025Abstract:Generating precise 3D molecular geometries is crucial for drug discovery and material science. While prior efforts leverage 1D representations like SELFIES to ensure molecular validity, they fail to fully exploit the rich chemical knowledge entangled within 1D models, leading to a disconnect between 1D syntactic generation and 3D geometric realization. To bridge this gap, we propose MolSculpt, a novel framework that "sculpts" 3D molecular geometries from chemical syntax. MolSculpt is built upon a frozen 1D molecular foundation model and a 3D molecular diffusion model. We introduce a set of learnable queries to extract inherent chemical knowledge from the foundation model, and a trainable projector then injects this cross-modal information into the conditioning space of the diffusion model to guide the 3D geometry generation. In this way, our model deeply integrates 1D latent chemical knowledge into the 3D generation process through end-to-end optimization. Experiments demonstrate that MolSculpt achieves state-of-the-art (SOTA) performance in \textit{de novo} 3D molecule generation and conditional 3D molecule generation, showing superior 3D fidelity and stability on both the GEOM-DRUGS and QM9 datasets. Code is available at https://github.com/SakuraTroyChen/MolSculpt.
Decouple Graph Neural Networks: Train Multiple Simple GNNs Simultaneously Instead of One
Apr 20, 2023



Abstract:Graph neural networks (GNN) suffer from severe inefficiency. It is mainly caused by the exponential growth of node dependency with the increase of layers. It extremely limits the application of stochastic optimization algorithms so that the training of GNN is usually time-consuming. To address this problem, we propose to decouple a multi-layer GNN as multiple simple modules for more efficient training, which is comprised of classical forward training (FT)and designed backward training (BT). Under the proposed framework, each module can be trained efficiently in FT by stochastic algorithms without distortion of graph information owing to its simplicity. To avoid the only unidirectional information delivery of FT and sufficiently train shallow modules with the deeper ones, we develop a backward training mechanism that makes the former modules perceive the latter modules. The backward training introduces the reversed information delivery into the decoupled modules as well as the forward information delivery. To investigate how the decoupling and greedy training affect the representational capacity, we theoretically prove that the error produced by linear modules will not accumulate on unsupervised tasks in most cases. The theoretical and experimental results show that the proposed framework is highly efficient with reasonable performance.
A deep convolutional neural network approach to single-particle recognition in cryo-electron microscopy
Dec 31, 2016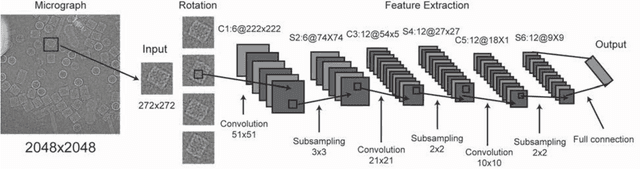
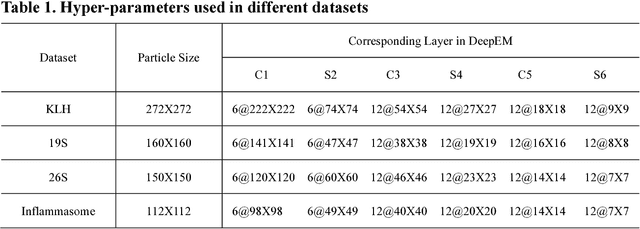
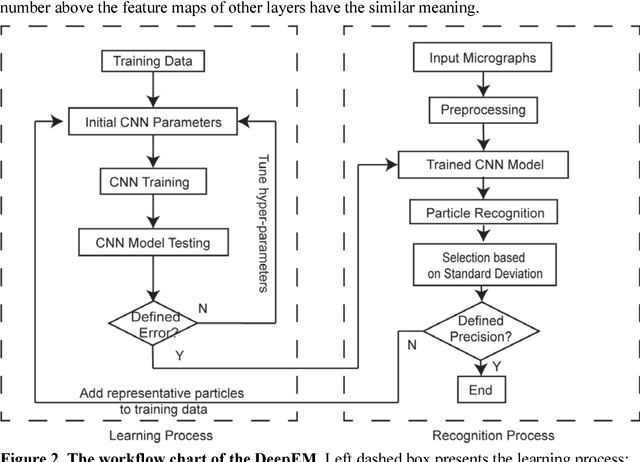
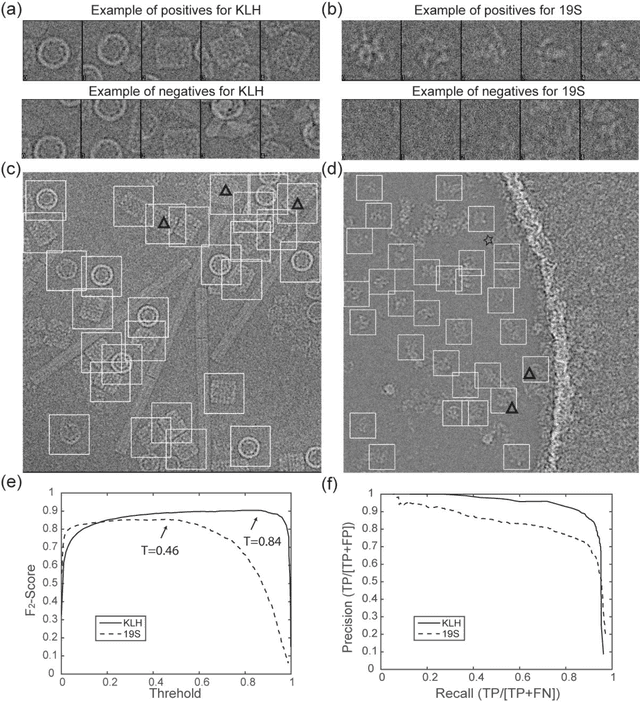
Abstract:Background: Single-particle cryo-electron microscopy (cryo-EM) has become a popular tool for structural determination of biological macromolecular complexes. High-resolution cryo-EM reconstruction often requires hundreds of thousands of single-particle images. Particle extraction from experimental micrographs thus can be laborious and presents a major practical bottleneck in cryo-EM structural determination. Existing computational methods of particle picking often use low-resolution templates as inputs for particle matching, making it possible to cause reference-dependent bias. It is critical to develop a highly efficient template-free method to automatically recognize particle images from cryo-EM micrographs. Results: We developed a deep learning-based algorithmic framework, DeepEM, for single-particle recognition from noisy cryo-EM micrographs, enabling automated particle picking, selection and verification in an integrated fashion. The kernel of DeepEM is built upon a convolutional neural network (CNN) of eight layers, which can be recursively trained to be highly "knowledgeable". Our approach exhibits improved performance and high precision when tested on the standard KLH dataset. Application of DeepEM to several challenging experimental cryo-EM datasets demonstrates its capability in avoiding selection of un-wanted particles and non-particles even when true particles contain fewer features. Conclusions: The DeepEM method derived from a deep CNN allows automated particle extraction from raw cryo-EM micrographs in the absence of templates, which demonstrated improved performance, objectivity and accuracy. Application of this novel approach is expected to free the labor involved in single-particle verification, thus promoting the efficiency of cryo-EM data processing.
* 26 pages, 6 figures, 1 table
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge