Vikas Garg
On topological descriptors for graph products
Nov 12, 2025Abstract:Topological descriptors have been increasingly utilized for capturing multiscale structural information in relational data. In this work, we consider various filtrations on the (box) product of graphs and the effect on their outputs on the topological descriptors - the Euler characteristic (EC) and persistent homology (PH). In particular, we establish a complete characterization of the expressive power of EC on general color-based filtrations. We also show that the PH descriptors of (virtual) graph products contain strictly more information than the computation on individual graphs, whereas EC does not. Additionally, we provide algorithms to compute the PH diagrams of the product of vertex- and edge-level filtrations on the graph product. We also substantiate our theoretical analysis with empirical investigations on runtime analysis, expressivity, and graph classification performance. Overall, this work paves way for powerful graph persistent descriptors via product filtrations. Code is available at https://github.com/Aalto-QuML/tda_graph_product.
Heat Kernel Goes Topological
Jul 16, 2025Abstract:Topological neural networks have emerged as powerful successors of graph neural networks. However, they typically involve higher-order message passing, which incurs significant computational expense. We circumvent this issue with a novel topological framework that introduces a Laplacian operator on combinatorial complexes (CCs), enabling efficient computation of heat kernels that serve as node descriptors. Our approach captures multiscale information and enables permutation-equivariant representations, allowing easy integration into modern transformer-based architectures. Theoretically, the proposed method is maximally expressive because it can distinguish arbitrary non-isomorphic CCs. Empirically, it significantly outperforms existing topological methods in terms of computational efficiency. Besides demonstrating competitive performance with the state-of-the-art descriptors on standard molecular datasets, it exhibits superior capability in distinguishing complex topological structures and avoiding blind spots on topological benchmarks. Overall, this work advances topological deep learning by providing expressive yet scalable representations, thereby opening up exciting avenues for molecular classification and property prediction tasks.
Graph Persistence goes Spectral
Jun 06, 2025Abstract:Including intricate topological information (e.g., cycles) provably enhances the expressivity of message-passing graph neural networks (GNNs) beyond the Weisfeiler-Leman (WL) hierarchy. Consequently, Persistent Homology (PH) methods are increasingly employed for graph representation learning. In this context, recent works have proposed decorating classical PH diagrams with vertex and edge features for improved expressivity. However, due to their dependence on features, these methods still fail to capture basic graph structural information. In this paper, we propose SpectRe -- a new topological descriptor for graphs that integrates spectral information into PH diagrams. Notably, SpectRe is strictly more expressive than existing descriptors on graphs. We also introduce notions of global and local stability to analyze existing descriptors and establish that SpectRe is locally stable. Finally, experiments on synthetic and real-world datasets demonstrate the effectiveness of SpectRe and its potential to enhance the capabilities of graph models in relevant learning tasks.
Positional Encoding meets Persistent Homology on Graphs
Jun 06, 2025Abstract:The local inductive bias of message-passing graph neural networks (GNNs) hampers their ability to exploit key structural information (e.g., connectivity and cycles). Positional encoding (PE) and Persistent Homology (PH) have emerged as two promising approaches to mitigate this issue. PE schemes endow GNNs with location-aware features, while PH methods enhance GNNs with multiresolution topological features. However, a rigorous theoretical characterization of the relative merits and shortcomings of PE and PH has remained elusive. We bridge this gap by establishing that neither paradigm is more expressive than the other, providing novel constructions where one approach fails but the other succeeds. Our insights inform the design of a novel learnable method, PiPE (Persistence-informed Positional Encoding), which is provably more expressive than both PH and PE. PiPE demonstrates strong performance across a variety of tasks (e.g., molecule property prediction, graph classification, and out-of-distribution generalization), thereby advancing the frontiers of graph representation learning. Code is available at https://github.com/Aalto-QuML/PIPE.
Spacetime Geometry of Denoising in Diffusion Models
May 23, 2025Abstract:We present a novel perspective on diffusion models using the framework of information geometry. We show that the set of noisy samples, taken across all noise levels simultaneously, forms a statistical manifold -- a family of denoising probability distributions. Interpreting the noise level as a temporal parameter, we refer to this manifold as spacetime. This manifold naturally carries a Fisher-Rao metric, which defines geodesics -- shortest paths between noisy points. Notably, this family of distributions is exponential, enabling efficient geodesic computation even in high-dimensional settings without retraining or fine-tuning. We demonstrate the practical value of this geometric viewpoint in transition path sampling, where spacetime geodesics define smooth sequences of Boltzmann distributions, enabling the generation of continuous trajectories between low-energy metastable states. Code is available at: https://github.com/Aalto-QuML/diffusion-spacetime-geometry.
Heterophily-informed Message Passing
Apr 28, 2025



Abstract:Graph neural networks (GNNs) are known to be vulnerable to oversmoothing due to their implicit homophily assumption. We mitigate this problem with a novel scheme that regulates the aggregation of messages, modulating the type and extent of message passing locally thereby preserving both the low and high-frequency components of information. Our approach relies solely on learnt embeddings, obviating the need for auxiliary labels, thus extending the benefits of heterophily-aware embeddings to broader applications, e.g., generative modelling. Our experiments, conducted across various data sets and GNN architectures, demonstrate performance enhancements and reveal heterophily patterns across standard classification benchmarks. Furthermore, application to molecular generation showcases notable performance improvements on chemoinformatics benchmarks.
Robust Simulation-Based Inference under Missing Data via Neural Processes
Mar 03, 2025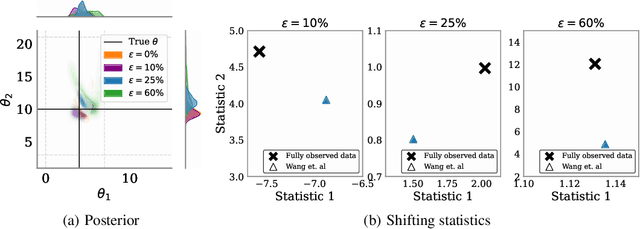

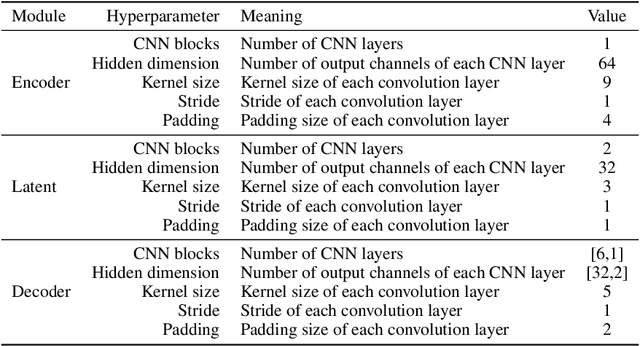
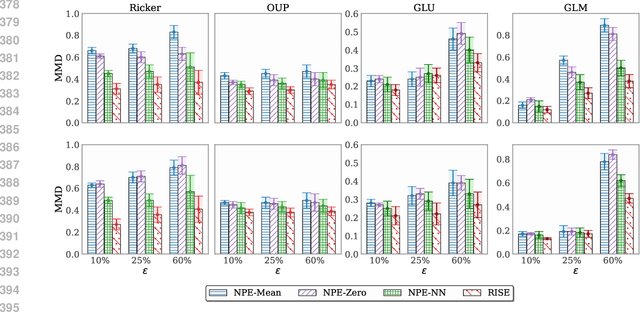
Abstract:Simulation-based inference (SBI) methods typically require fully observed data to infer parameters of models with intractable likelihood functions. However, datasets often contain missing values due to incomplete observations, data corruptions (common in astrophysics), or instrument limitations (e.g., in high-energy physics applications). In such scenarios, missing data must be imputed before applying any SBI method. We formalize the problem of missing data in SBI and demonstrate that naive imputation methods can introduce bias in the estimation of SBI posterior. We also introduce a novel amortized method that addresses this issue by jointly learning the imputation model and the inference network within a neural posterior estimation (NPE) framework. Extensive empirical results on SBI benchmarks show that our approach provides robust inference outcomes compared to standard baselines for varying levels of missing data. Moreover, we demonstrate the merits of our imputation model on two real-world bioactivity datasets (Adrenergic and Kinase assays). Code is available at https://github.com/Aalto-QuML/RISE.
Devil is in the Details: Density Guidance for Detail-Aware Generation with Flow Models
Feb 09, 2025



Abstract:Diffusion models have emerged as a powerful class of generative models, capable of producing high-quality images by mapping noise to a data distribution. However, recent findings suggest that image likelihood does not align with perceptual quality: high-likelihood samples tend to be smooth, while lower-likelihood ones are more detailed. Controlling sample density is thus crucial for balancing realism and detail. In this paper, we analyze an existing technique, Prior Guidance, which scales the latent code to influence image detail. We introduce score alignment, a condition that explains why this method works and show that it can be tractably checked for any continuous normalizing flow model. We then propose Density Guidance, a principled modification of the generative ODE that enables exact log-density control during sampling. Finally, we extend Density Guidance to stochastic sampling, ensuring precise log-density control while allowing controlled variation in structure or fine details. Our experiments demonstrate that these techniques provide fine-grained control over image detail without compromising sample quality.
Diffusion Models as Cartoonists! The Curious Case of High Density Regions
Nov 02, 2024Abstract:We investigate what kind of images lie in the high-density regions of diffusion models. We introduce a theoretical mode-tracking process capable of pinpointing the exact mode of the denoising distribution, and we propose a practical high-probability sampler that consistently generates images of higher likelihood than usual samplers. Our empirical findings reveal the existence of significantly higher likelihood samples that typical samplers do not produce, often manifesting as cartoon-like drawings or blurry images depending on the noise level. Curiously, these patterns emerge in datasets devoid of such examples. We also present a novel approach to track sample likelihoods in diffusion SDEs, which remarkably incurs no additional computational cost.
Diffusion Twigs with Loop Guidance for Conditional Graph Generation
Oct 31, 2024



Abstract:We introduce a novel score-based diffusion framework named Twigs that incorporates multiple co-evolving flows for enriching conditional generation tasks. Specifically, a central or trunk diffusion process is associated with a primary variable (e.g., graph structure), and additional offshoot or stem processes are dedicated to dependent variables (e.g., graph properties or labels). A new strategy, which we call loop guidance, effectively orchestrates the flow of information between the trunk and the stem processes during sampling. This approach allows us to uncover intricate interactions and dependencies, and unlock new generative capabilities. We provide extensive experiments to demonstrate strong performance gains of the proposed method over contemporary baselines in the context of conditional graph generation, underscoring the potential of Twigs in challenging generative tasks such as inverse molecular design and molecular optimization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge