Tonio Ball
Deep learning with convolutional neural networks for EEG decoding and visualization
Jun 08, 2018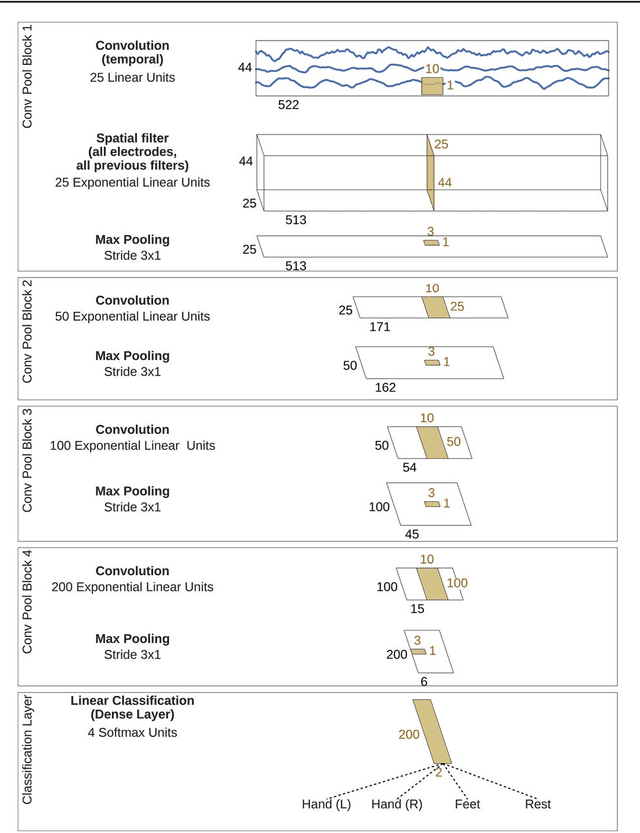
Abstract:PLEASE READ AND CITE THE REVISED VERSION at Human Brain Mapping: http://onlinelibrary.wiley.com/doi/10.1002/hbm.23730/full Code available here: https://github.com/robintibor/braindecode
EEG-GAN: Generative adversarial networks for electroencephalograhic (EEG) brain signals
Jun 05, 2018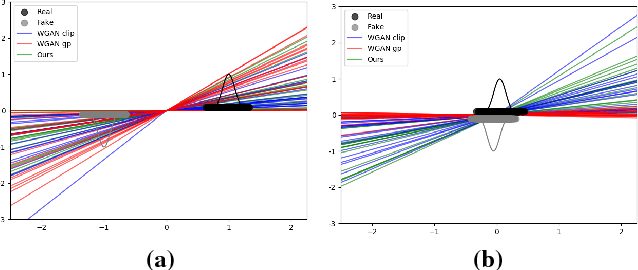
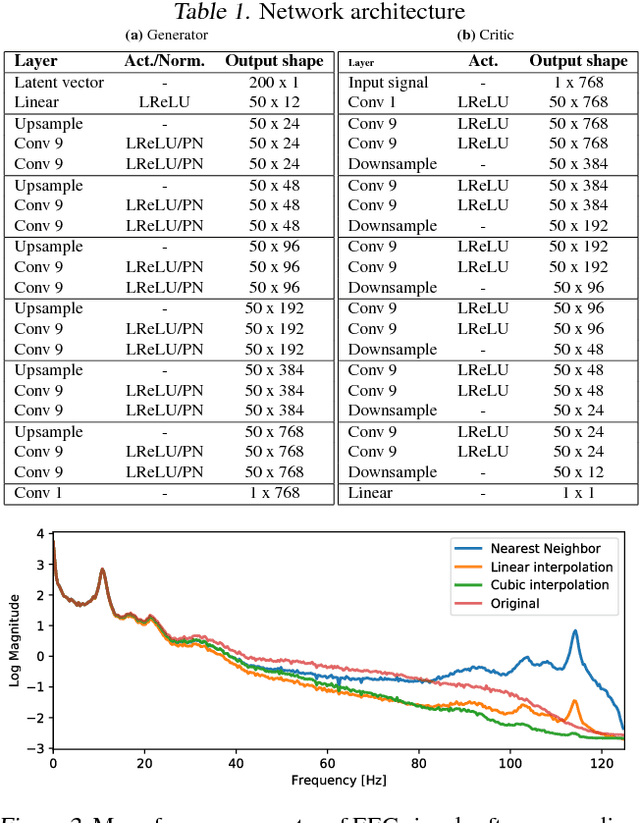
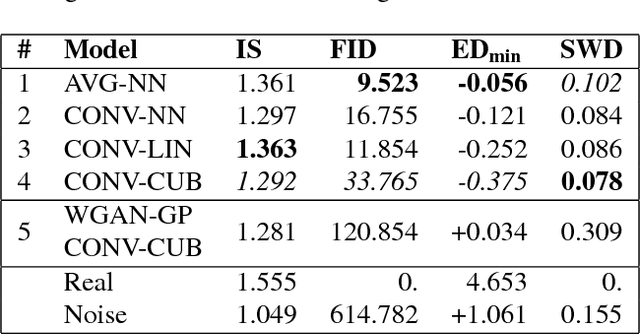
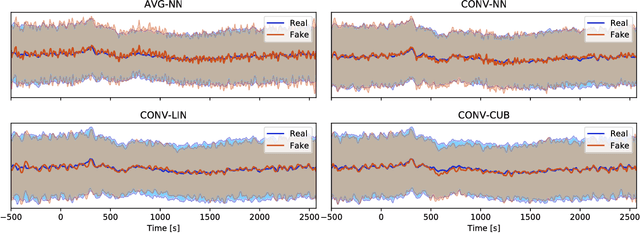
Abstract:Generative adversarial networks (GANs) are recently highly successful in generative applications involving images and start being applied to time series data. Here we describe EEG-GAN as a framework to generate electroencephalographic (EEG) brain signals. We introduce a modification to the improved training of Wasserstein GANs to stabilize training and investigate a range of architectural choices critical for time series generation (most notably up- and down-sampling). For evaluation we consider and compare different metrics such as Inception score, Frechet inception distance and sliced Wasserstein distance, together showing that our EEG-GAN framework generated naturalistic EEG examples. It thus opens up a range of new generative application scenarios in the neuroscientific and neurological context, such as data augmentation in brain-computer interfacing tasks, EEG super-sampling, or restoration of corrupted data segments. The possibility to generate signals of a certain class and/or with specific properties may also open a new avenue for research into the underlying structure of brain signals.
Deep learning with convolutional neural networks for decoding and visualization of EEG pathology
Jan 11, 2018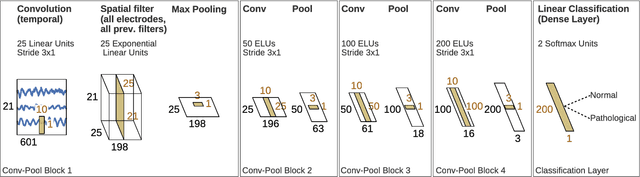
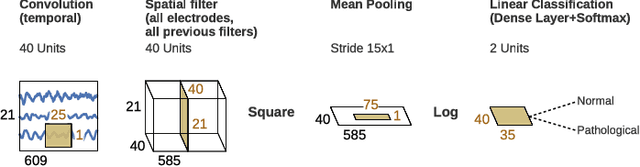
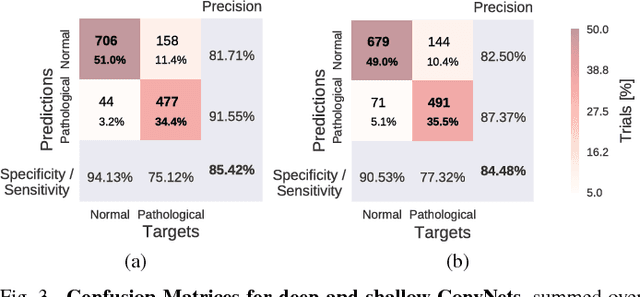
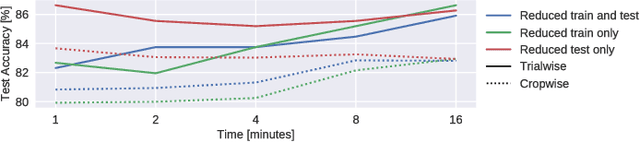
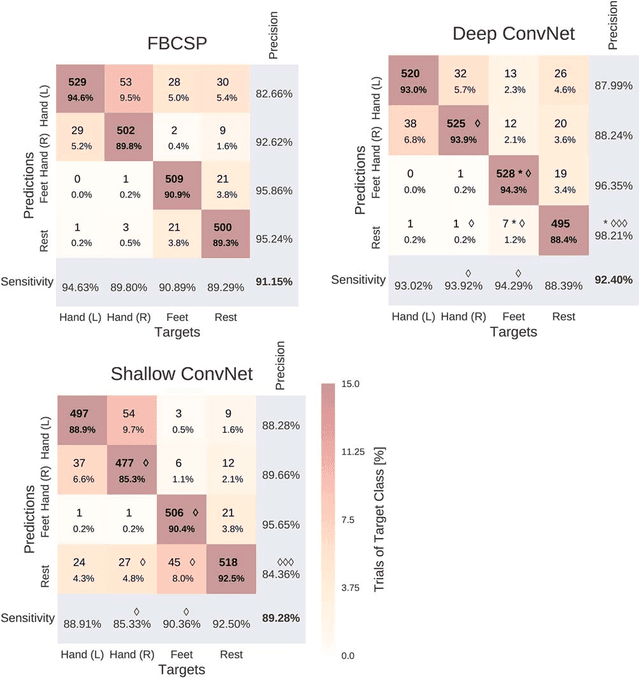
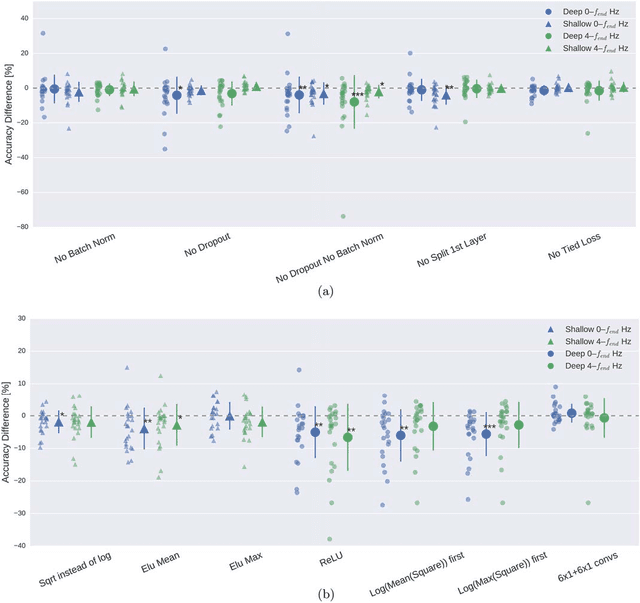
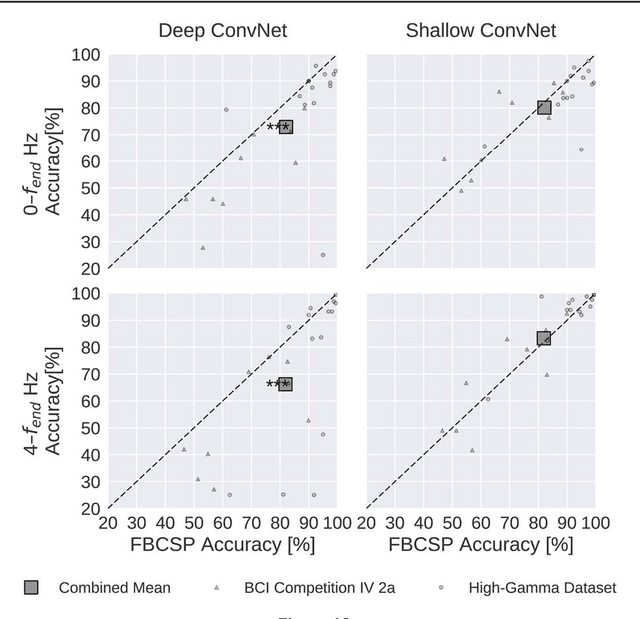
Abstract:We apply convolutional neural networks (ConvNets) to the task of distinguishing pathological from normal EEG recordings in the Temple University Hospital EEG Abnormal Corpus. We use two basic, shallow and deep ConvNet architectures recently shown to decode task-related information from EEG at least as well as established algorithms designed for this purpose. In decoding EEG pathology, both ConvNets reached substantially better accuracies (about 6% better, ~85% vs. ~79%) than the only published result for this dataset, and were still better when using only 1 minute of each recording for training and only six seconds of each recording for testing. We used automated methods to optimize architectural hyperparameters and found intriguingly different ConvNet architectures, e.g., with max pooling as the only nonlinearity. Visualizations of the ConvNet decoding behavior showed that they used spectral power changes in the delta (0-4 Hz) and theta (4-8 Hz) frequency range, possibly alongside other features, consistent with expectations derived from spectral analysis of the EEG data and from the textual medical reports. Analysis of the textual medical reports also highlighted the potential for accuracy increases by integrating contextual information, such as the age of subjects. In summary, the ConvNets and visualization techniques used in this study constitute a next step towards clinically useful automated EEG diagnosis and establish a new baseline for future work on this topic.
Deep Transfer Learning for Error Decoding from Non-Invasive EEG
Jan 10, 2018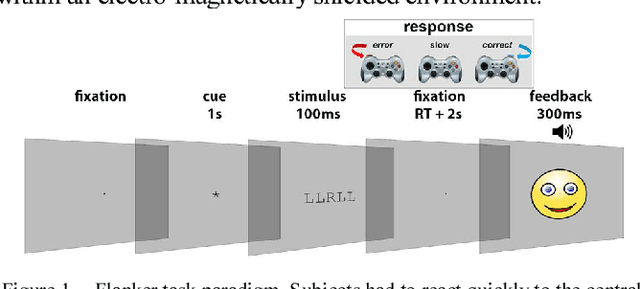
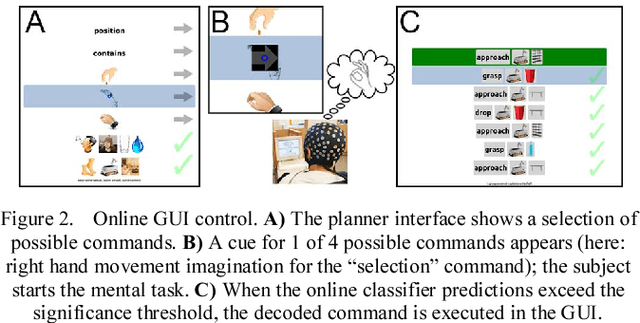
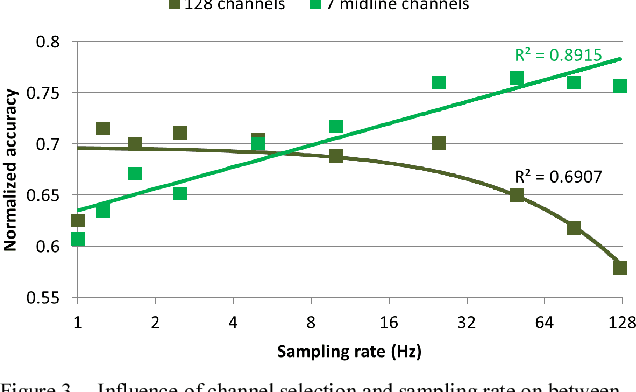
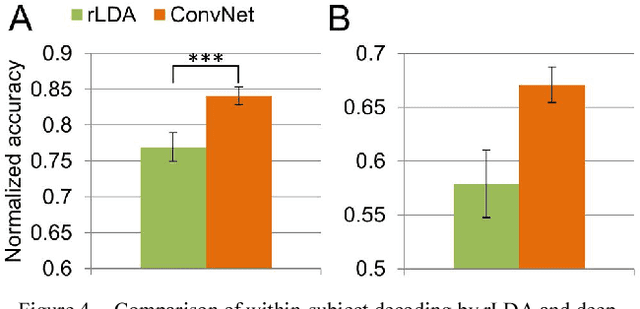
Abstract:We recorded high-density EEG in a flanker task experiment (31 subjects) and an online BCI control paradigm (4 subjects). On these datasets, we evaluated the use of transfer learning for error decoding with deep convolutional neural networks (deep ConvNets). In comparison with a regularized linear discriminant analysis (rLDA) classifier, ConvNets were significantly better in both intra- and inter-subject decoding, achieving an average accuracy of 84.1 % within subject and 81.7 % on unknown subjects (flanker task). Neither method was, however, able to generalize reliably between paradigms. Visualization of features the ConvNets learned from the data showed plausible patterns of brain activity, revealing both similarities and differences between the different kinds of errors. Our findings indicate that deep learning techniques are useful to infer information about the correctness of action in BCI applications, particularly for the transfer of pre-trained classifiers to new recording sessions or subjects.
Hierarchical internal representation of spectral features in deep convolutional networks trained for EEG decoding
Dec 15, 2017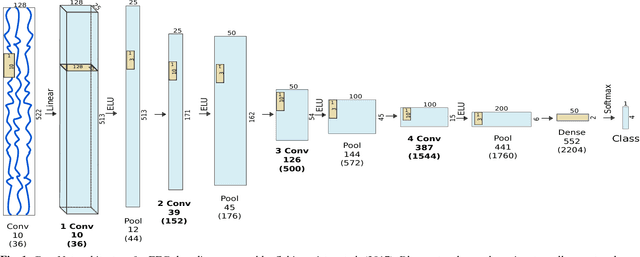
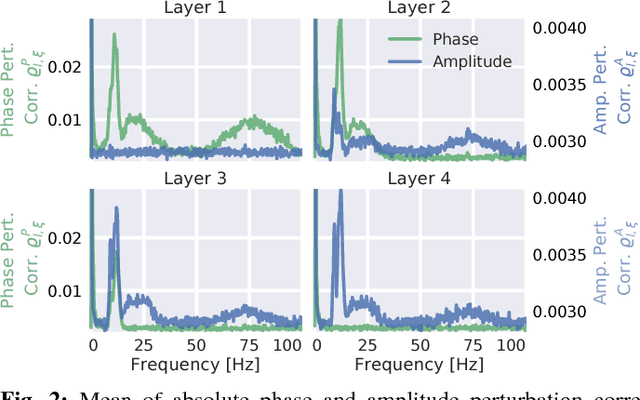
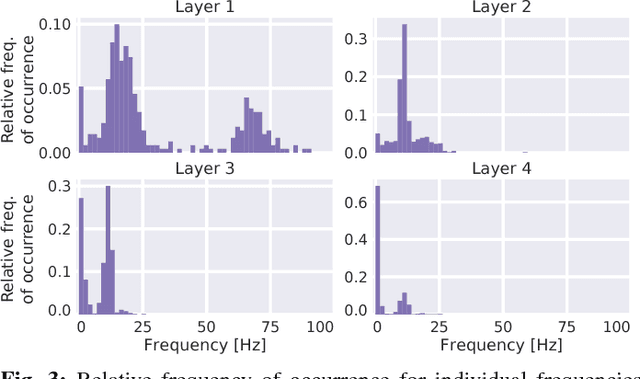
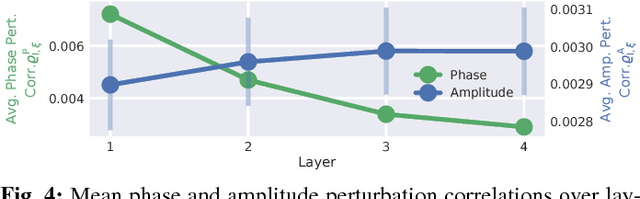
Abstract:Recently, there is increasing interest and research on the interpretability of machine learning models, for example how they transform and internally represent EEG signals in Brain-Computer Interface (BCI) applications. This can help to understand the limits of the model and how it may be improved, in addition to possibly provide insight about the data itself. Schirrmeister et al. (2017) have recently reported promising results for EEG decoding with deep convolutional neural networks (ConvNets) trained in an end-to-end manner and, with a causal visualization approach, showed that they learn to use spectral amplitude changes in the input. In this study, we investigate how ConvNets represent spectral features through the sequence of intermediate stages of the network. We show higher sensitivity to EEG phase features at earlier stages and higher sensitivity to EEG amplitude features at later stages. Intriguingly, we observed a specialization of individual stages of the network to the classical EEG frequency bands alpha, beta, and high gamma. Furthermore, we find first evidence that particularly in the last convolutional layer, the network learns to detect more complex oscillatory patterns beyond spectral phase and amplitude, reminiscent of the representation of complex visual features in later layers of ConvNets in computer vision tasks. Our findings thus provide insights into how ConvNets hierarchically represent spectral EEG features in their intermediate layers and suggest that ConvNets can exploit and might help to better understand the compositional structure of EEG time series.
The signature of robot action success in EEG signals of a human observer: Decoding and visualization using deep convolutional neural networks
Nov 16, 2017

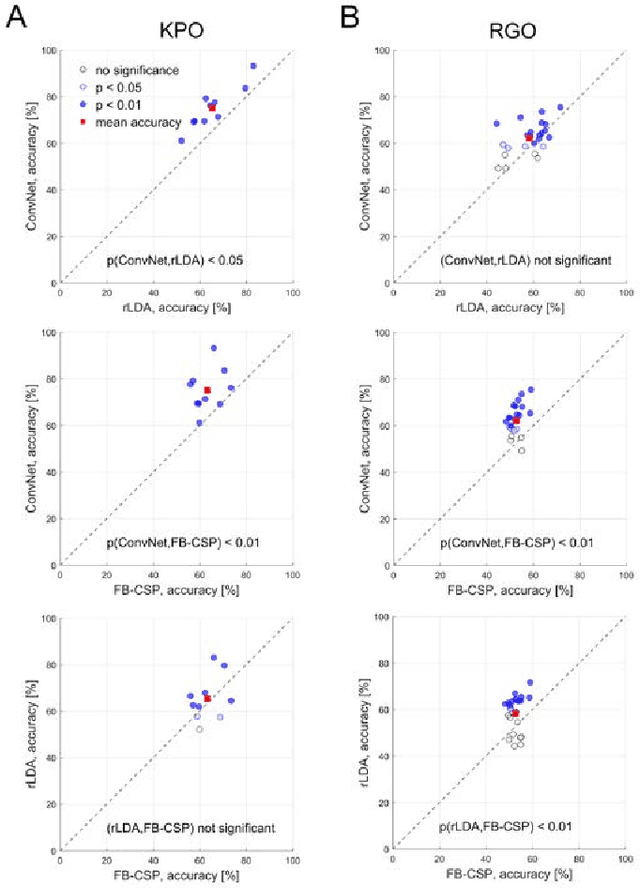
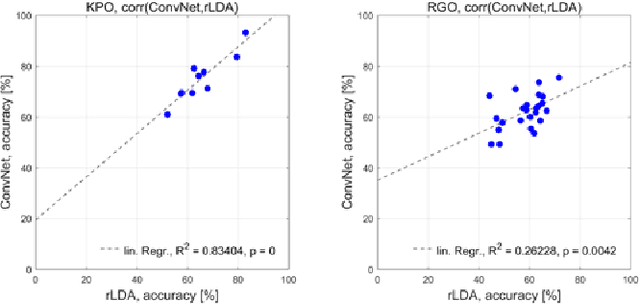
Abstract:The importance of robotic assistive devices grows in our work and everyday life. Cooperative scenarios involving both robots and humans require safe human-robot interaction. One important aspect here is the management of robot errors, including fast and accurate online robot-error detection and correction. Analysis of brain signals from a human interacting with a robot may help identifying robot errors, but accuracies of such analyses have still substantial space for improvement. In this paper we evaluate whether a novel framework based on deep convolutional neural networks (deep ConvNets) could improve the accuracy of decoding robot errors from the EEG of a human observer, both during an object grasping and a pouring task. We show that deep ConvNets reached significantly higher accuracies than both regularized Linear Discriminant Analysis (rLDA) and filter bank common spatial patterns (FB-CSP) combined with rLDA, both widely used EEG classifiers. Deep ConvNets reached mean accuracies of 75% +/- 9 %, rLDA 65% +/- 10% and FB-CSP + rLDA 63% +/- 6% for decoding of erroneous vs. correct trials. Visualization of the time-domain EEG features learned by the ConvNets to decode errors revealed spatiotemporal patterns that reflected differences between the two experimental paradigms. Across subjects, ConvNet decoding accuracies were significantly correlated with those obtained with rLDA, but not CSP, indicating that in the present context ConvNets behaved more 'rLDA-like' (but consistently better), while in a previous decoding study with another task but the same ConvNet architecture, it was found to behave more 'CSP-like'. Our findings thus provide further support for the assumption that deep ConvNets are a versatile addition to the existing toolbox of EEG decoding techniques, and we discuss steps how ConvNet EEG decoding performance could be further optimized.
Brain Responses During Robot-Error Observation
Aug 16, 2017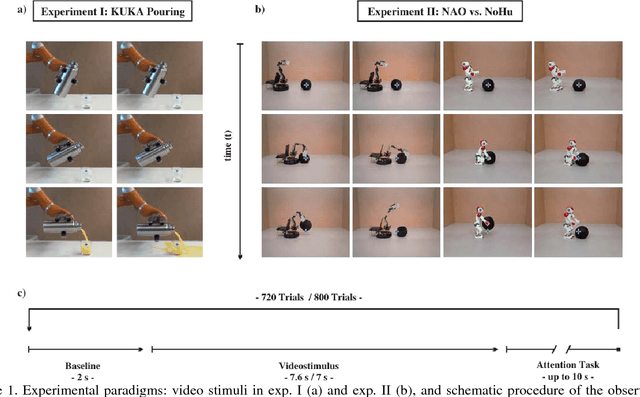
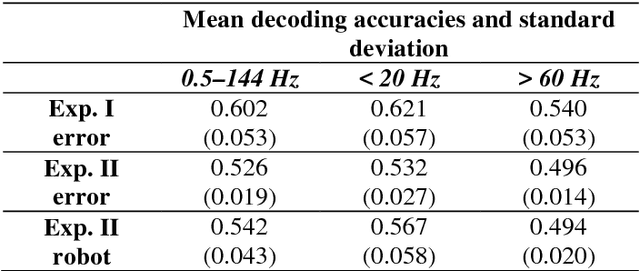
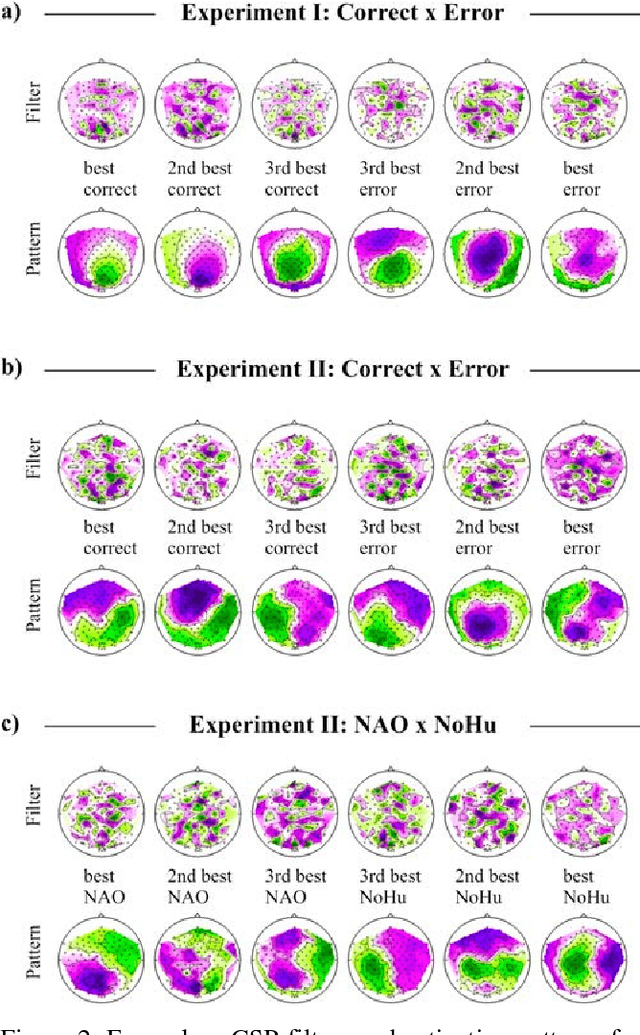
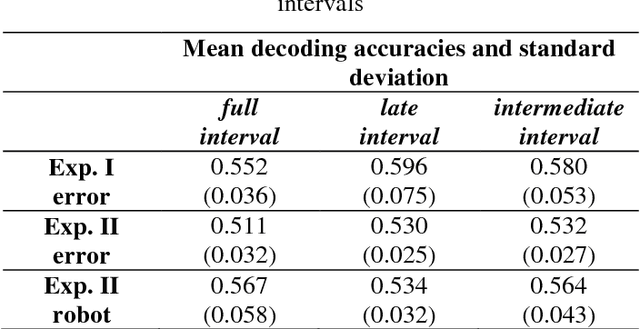
Abstract:Brain-controlled robots are a promising new type of assistive device for severely impaired persons. Little is however known about how to optimize the interaction of humans and brain-controlled robots. Information about the human's perceived correctness of robot performance might provide a useful teaching signal for adaptive control algorithms and thus help enhancing robot control. Here, we studied whether watching robots perform erroneous vs. correct action elicits differential brain responses that can be decoded from single trials of electroencephalographic (EEG) recordings, and whether brain activity during human-robot interaction is modulated by the robot's visual similarity to a human. To address these topics, we designed two experiments. In experiment I, participants watched a robot arm pour liquid into a cup. The robot performed the action either erroneously or correctly, i.e. it either spilled some liquid or not. In experiment II, participants observed two different types of robots, humanoid and non-humanoid, grabbing a ball. The robots either managed to grab the ball or not. We recorded high-resolution EEG during the observation tasks in both experiments to train a Filter Bank Common Spatial Pattern (FBCSP) pipeline on the multivariate EEG signal and decode for the correctness of the observed action, and for the type of the observed robot. Our findings show that it was possible to decode both correctness and robot type for the majority of participants significantly, although often just slightly, above chance level. Our findings suggest that non-invasive recordings of brain responses elicited when observing robots indeed contain decodable information about the correctness of the robot's action and the type of observed robot.
Causal and anti-causal learning in pattern recognition for neuroimaging
Dec 15, 2015
Abstract:Pattern recognition in neuroimaging distinguishes between two types of models: encoding- and decoding models. This distinction is based on the insight that brain state features, that are found to be relevant in an experimental paradigm, carry a different meaning in encoding- than in decoding models. In this paper, we argue that this distinction is not sufficient: Relevant features in encoding- and decoding models carry a different meaning depending on whether they represent causal- or anti-causal relations. We provide a theoretical justification for this argument and conclude that causal inference is essential for interpretation in neuroimaging.
* accepted manuscript
Decoding index finger position from EEG using random forests
Dec 14, 2015
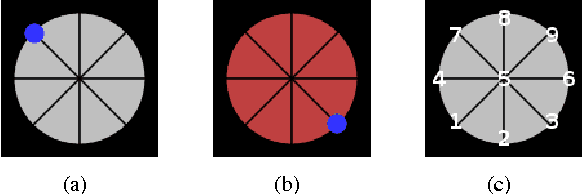
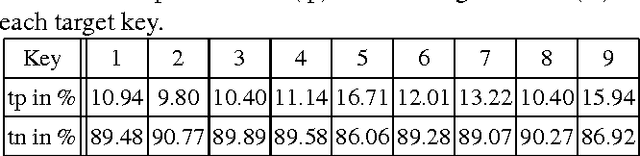
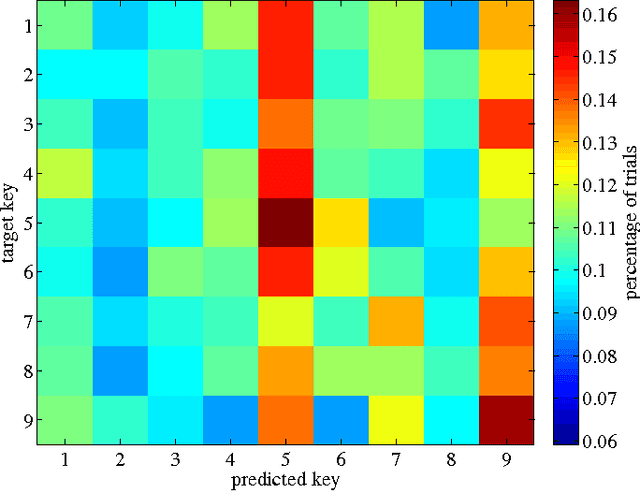
Abstract:While invasively recorded brain activity is known to provide detailed information on motor commands, it is an open question at what level of detail information about positions of body parts can be decoded from non-invasively acquired signals. In this work it is shown that index finger positions can be differentiated from non-invasive electroencephalographic (EEG) recordings in healthy human subjects. Using a leave-one-subject-out cross-validation procedure, a random forest distinguished different index finger positions on a numerical keyboard above chance-level accuracy. Among the different spectral features investigated, high $\beta$-power (20-30 Hz) over contralateral sensorimotor cortex carried most information about finger position. Thus, these findings indicate that finger position is in principle decodable from non-invasive features of brain activity that generalize across individuals.
* accepted manuscript
Causal interpretation rules for encoding and decoding models in neuroimaging
Nov 15, 2015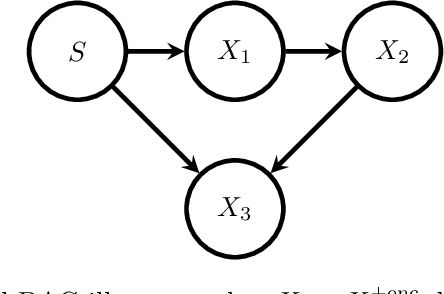
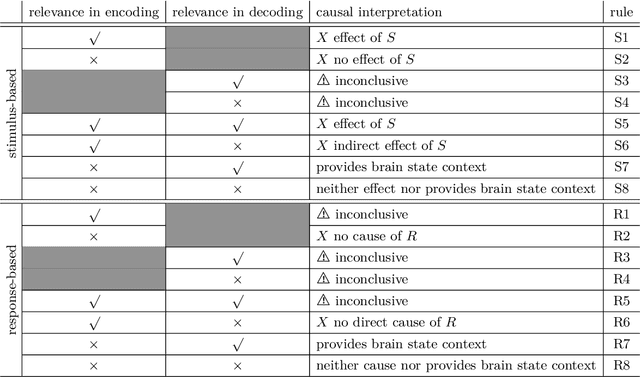

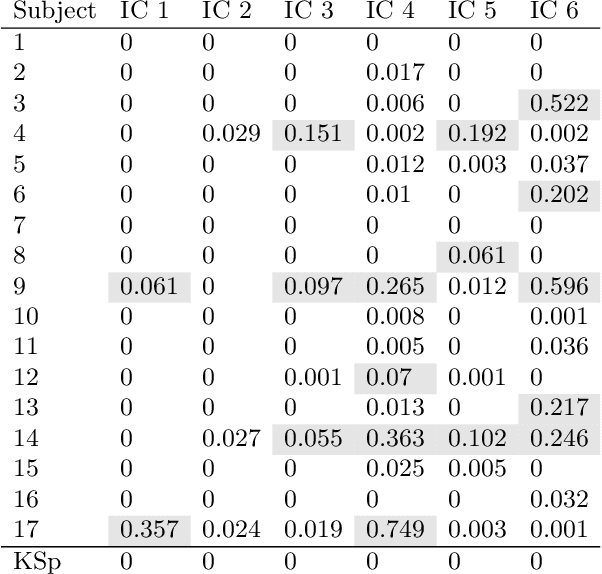
Abstract:Causal terminology is often introduced in the interpretation of encoding and decoding models trained on neuroimaging data. In this article, we investigate which causal statements are warranted and which ones are not supported by empirical evidence. We argue that the distinction between encoding and decoding models is not sufficient for this purpose: relevant features in encoding and decoding models carry a different meaning in stimulus- and in response-based experimental paradigms. We show that only encoding models in the stimulus-based setting support unambiguous causal interpretations. By combining encoding and decoding models trained on the same data, however, we obtain insights into causal relations beyond those that are implied by each individual model type. We illustrate the empirical relevance of our theoretical findings on EEG data recorded during a visuo-motor learning task.
* accepted manuscript
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge