Timothy Miller
Identifying Task Groupings for Multi-Task Learning Using Pointwise V-Usable Information
Oct 16, 2024Abstract:The success of multi-task learning can depend heavily on which tasks are grouped together. Naively grouping all tasks or a random set of tasks can result in negative transfer, with the multi-task models performing worse than single-task models. Though many efforts have been made to identify task groupings and to measure the relatedness among different tasks, it remains a challenging research topic to define a metric to identify the best task grouping out of a pool of many potential task combinations. We propose a metric of task relatedness based on task difficulty measured by pointwise V-usable information (PVI). PVI is a recently proposed metric to estimate how much usable information a dataset contains given a model. We hypothesize that tasks with not statistically different PVI estimates are similar enough to benefit from the joint learning process. We conduct comprehensive experiments to evaluate the feasibility of this metric for task grouping on 15 NLP datasets in the general, biomedical, and clinical domains. We compare the results of the joint learners against single learners, existing baseline methods, and recent large language models, including Llama 2 and GPT-4. The results show that by grouping tasks with similar PVI estimates, the joint learners yielded competitive results with fewer total parameters, with consistent performance across domains.
Improving Clinical NLP Performance through Language Model-Generated Synthetic Clinical Data
Mar 28, 2024
Abstract:Generative models have been showing potential for producing data in mass. This study explores the enhancement of clinical natural language processing performance by utilizing synthetic data generated from advanced language models. Promising results show feasible applications in such a high-stakes domain.
The impact of using an AI chatbot to respond to patient messages
Oct 26, 2023



Abstract:Documentation burden is a major contributor to clinician burnout, which is rising nationally and is an urgent threat to our ability to care for patients. Artificial intelligence (AI) chatbots, such as ChatGPT, could reduce clinician burden by assisting with documentation. Although many hospitals are actively integrating such systems into electronic medical record systems, AI chatbots utility and impact on clinical decision-making have not been studied for this intended use. We are the first to examine the utility of large language models in assisting clinicians draft responses to patient questions. In our two-stage cross-sectional study, 6 oncologists responded to 100 realistic synthetic cancer patient scenarios and portal messages developed to reflect common medical situations, first manually, then with AI assistance. We find AI-assisted responses were longer, less readable, but provided acceptable drafts without edits 58% of time. AI assistance improved efficiency 77% of time, with low harm risk (82% safe). However, 7.7% unedited AI responses could severely harm. In 31% cases, physicians thought AI drafts were human-written. AI assistance led to more patient education recommendations, fewer clinical actions than manual responses. Results show promise for AI to improve clinician efficiency and patient care through assisting documentation, if used judiciously. Monitoring model outputs and human-AI interaction remains crucial for safe implementation.
Leveraging A Medical Knowledge Graph into Large Language Models for Diagnosis Prediction
Aug 28, 2023



Abstract:Electronic Health Records (EHRs) and routine documentation practices play a vital role in patients' daily care, providing a holistic record of health, diagnoses, and treatment. However, complex and verbose EHR narratives overload healthcare providers, risking diagnostic inaccuracies. While Large Language Models (LLMs) have showcased their potential in diverse language tasks, their application in the healthcare arena needs to ensure the minimization of diagnostic errors and the prevention of patient harm. In this paper, we outline an innovative approach for augmenting the proficiency of LLMs in the realm of automated diagnosis generation, achieved through the incorporation of a medical knowledge graph (KG) and a novel graph model: Dr.Knows, inspired by the clinical diagnostic reasoning process. We derive the KG from the National Library of Medicine's Unified Medical Language System (UMLS), a robust repository of biomedical knowledge. Our method negates the need for pre-training and instead leverages the KG as an auxiliary instrument aiding in the interpretation and summarization of complex medical concepts. Using real-world hospital datasets, our experimental results demonstrate that the proposed approach of combining LLMs with KG has the potential to improve the accuracy of automated diagnosis generation. More importantly, our approach offers an explainable diagnostic pathway, edging us closer to the realization of AI-augmented diagnostic decision support systems.
Multi-Task Training with In-Domain Language Models for Diagnostic Reasoning
Jun 13, 2023Abstract:Generative artificial intelligence (AI) is a promising direction for augmenting clinical diagnostic decision support and reducing diagnostic errors, a leading contributor to medical errors. To further the development of clinical AI systems, the Diagnostic Reasoning Benchmark (DR.BENCH) was introduced as a comprehensive generative AI framework, comprised of six tasks representing key components in clinical reasoning. We present a comparative analysis of in-domain versus out-of-domain language models as well as multi-task versus single task training with a focus on the problem summarization task in DR.BENCH (Gao et al., 2023). We demonstrate that a multi-task, clinically trained language model outperforms its general domain counterpart by a large margin, establishing a new state-of-the-art performance, with a ROUGE-L score of 28.55. This research underscores the value of domain-specific training for optimizing clinical diagnostic reasoning tasks.
Overview of the Problem List Summarization (ProbSum) 2023 Shared Task on Summarizing Patients' Active Diagnoses and Problems from Electronic Health Record Progress Notes
Jun 08, 2023Abstract:The BioNLP Workshop 2023 initiated the launch of a shared task on Problem List Summarization (ProbSum) in January 2023. The aim of this shared task is to attract future research efforts in building NLP models for real-world diagnostic decision support applications, where a system generating relevant and accurate diagnoses will augment the healthcare providers decision-making process and improve the quality of care for patients. The goal for participants is to develop models that generated a list of diagnoses and problems using input from the daily care notes collected from the hospitalization of critically ill patients. Eight teams submitted their final systems to the shared task leaderboard. In this paper, we describe the tasks, datasets, evaluation metrics, and baseline systems. Additionally, the techniques and results of the evaluation of the different approaches tried by the participating teams are summarized.
Progress Note Understanding -- Assessment and Plan Reasoning: Overview of the 2022 N2C2 Track 3 Shared Task
Mar 14, 2023



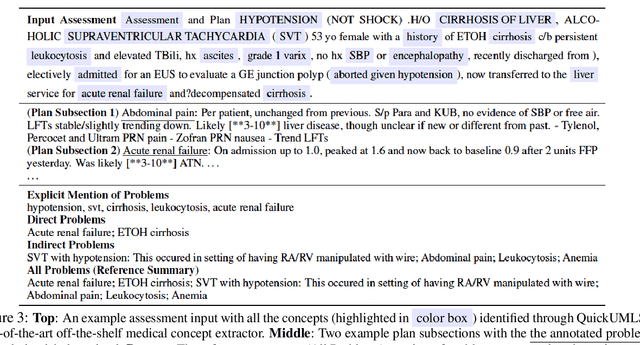
Abstract:Daily progress notes are common types in the electronic health record (EHR) where healthcare providers document the patient's daily progress and treatment plans. The EHR is designed to document all the care provided to patients, but it also enables note bloat with extraneous information that distracts from the diagnoses and treatment plans. Applications of natural language processing (NLP) in the EHR is a growing field with the majority of methods in information extraction. Few tasks use NLP methods for downstream diagnostic decision support. We introduced the 2022 National NLP Clinical Challenge (N2C2) Track 3: Progress Note Understanding - Assessment and Plan Reasoning as one step towards a new suite of tasks. The Assessment and Plan Reasoning task focuses on the most critical components of progress notes, Assessment and Plan subsections where health problems and diagnoses are contained. The goal of the task was to develop and evaluate NLP systems that automatically predict causal relations between the overall status of the patient contained in the Assessment section and its relation to each component of the Plan section which contains the diagnoses and treatment plans. The goal of the task was to identify and prioritize diagnoses as the first steps in diagnostic decision support to find the most relevant information in long documents like daily progress notes. We present the results of 2022 n2c2 Track 3 and provide a description of the data, evaluation, participation and system performance.
DR.BENCH: Diagnostic Reasoning Benchmark for Clinical Natural Language Processing
Sep 29, 2022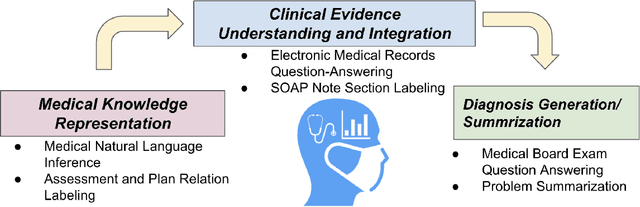
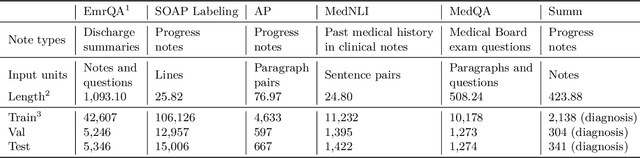
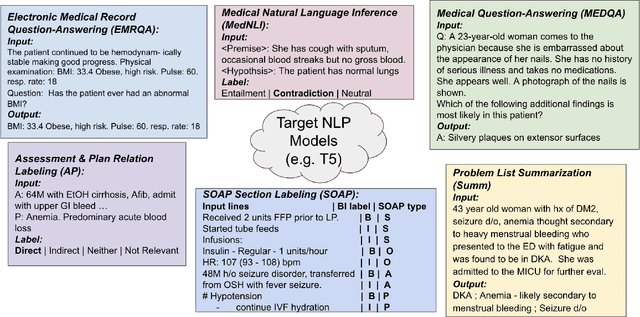
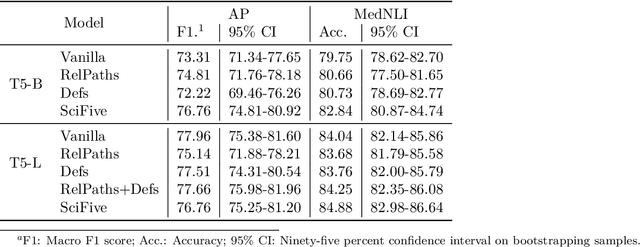
Abstract:The meaningful use of electronic health records (EHR) continues to progress in the digital era with clinical decision support systems augmented by artificial intelligence. A priority in improving provider experience is to overcome information overload and reduce the cognitive burden so fewer medical errors and cognitive biases are introduced during patient care. One major type of medical error is diagnostic error due to systematic or predictable errors in judgment that rely on heuristics. The potential for clinical natural language processing (cNLP) to model diagnostic reasoning in humans with forward reasoning from data to diagnosis and potentially reduce the cognitive burden and medical error has not been investigated. Existing tasks to advance the science in cNLP have largely focused on information extraction and named entity recognition through classification tasks. We introduce a novel suite of tasks coined as Diagnostic Reasoning Benchmarks, DR.BENCH, as a new benchmark for developing and evaluating cNLP models with clinical diagnostic reasoning ability. The suite includes six tasks from ten publicly available datasets addressing clinical text understanding, medical knowledge reasoning, and diagnosis generation. DR.BENCH is the first clinical suite of tasks designed to be a natural language generation framework to evaluate pre-trained language models. Experiments with state-of-the-art pre-trained generative language models using large general domain models and models that were continually trained on a medical corpus demonstrate opportunities for improvement when evaluated in DR. BENCH. We share DR. BENCH as a publicly available GitLab repository with a systematic approach to load and evaluate models for the cNLP community.
Summarizing Patients Problems from Hospital Progress Notes Using Pre-trained Sequence-to-Sequence Models
Aug 17, 2022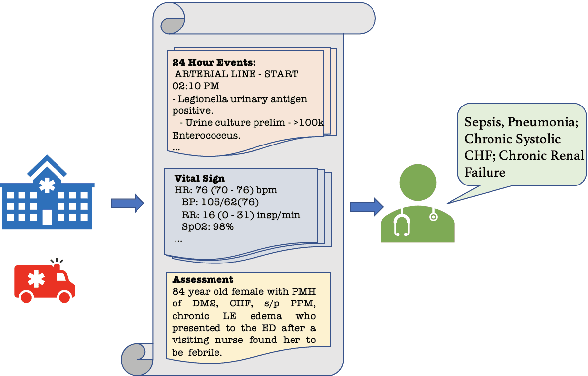
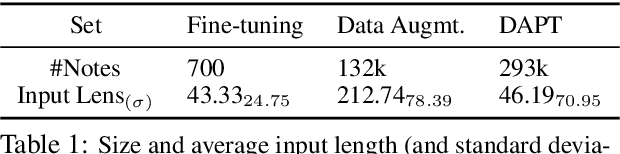
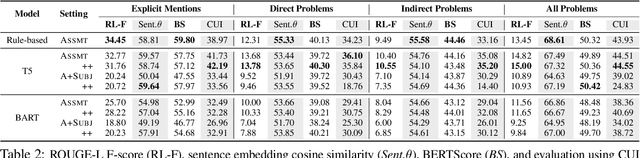
Abstract:Automatically summarizing patients' main problems from daily progress notes using natural language processing methods helps to battle against information and cognitive overload in hospital settings and potentially assists providers with computerized diagnostic decision support. Problem list summarization requires a model to understand, abstract, and generate clinical documentation. In this work, we propose a new NLP task that aims to generate a list of problems in a patient's daily care plan using input from the provider's progress notes during hospitalization. We investigate the performance of T5 and BART, two state-of-the-art seq2seq transformer architectures, in solving this problem. We provide a corpus built on top of progress notes from publicly available electronic health record progress notes in the Medical Information Mart for Intensive Care (MIMIC)-III. T5 and BART are trained on general domain text, and we experiment with a data augmentation method and a domain adaptation pre-training method to increase exposure to medical vocabulary and knowledge. Evaluation methods include ROUGE, BERTScore, cosine similarity on sentence embedding, and F-score on medical concepts. Results show that T5 with domain adaptive pre-training achieves significant performance gains compared to a rule-based system and general domain pre-trained language models, indicating a promising direction for tackling the problem summarization task.
Hierarchical Annotation for Building A Suite of Clinical Natural Language Processing Tasks: Progress Note Understanding
Apr 06, 2022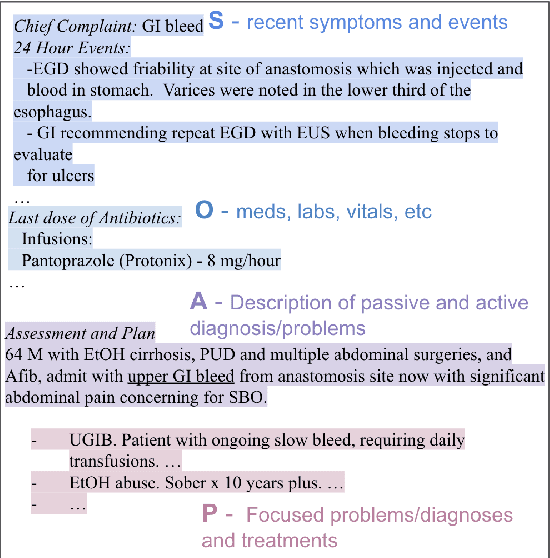
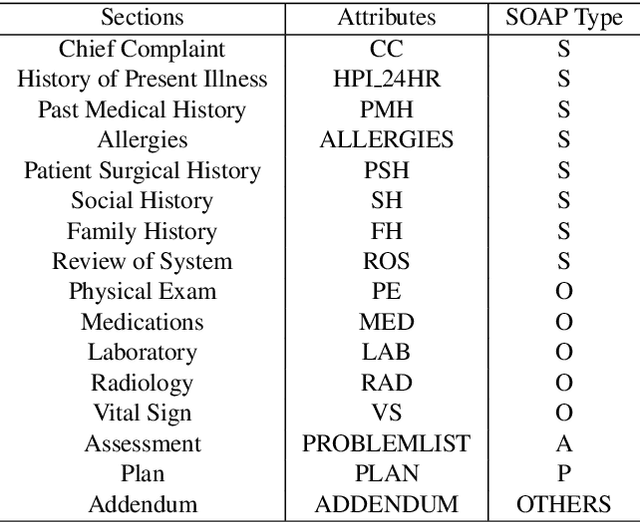
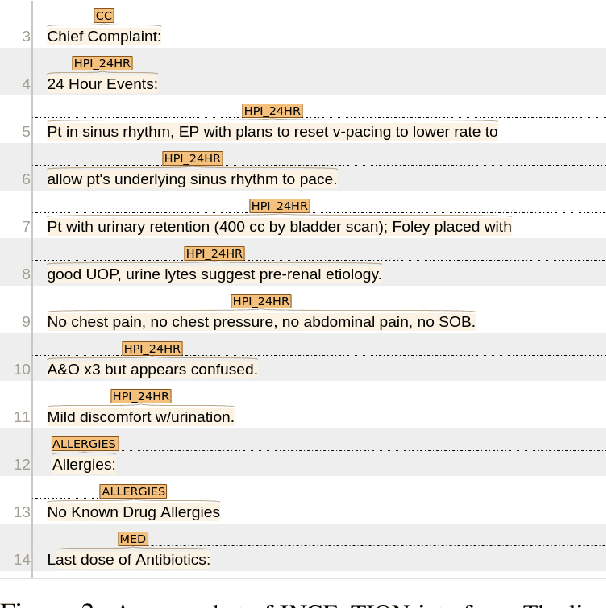
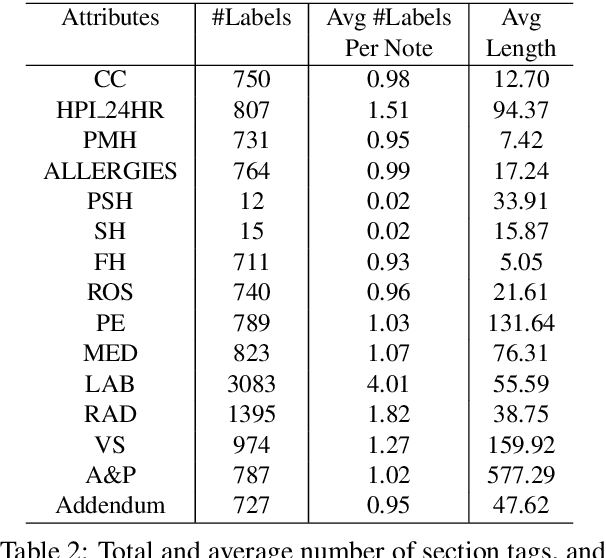
Abstract:Applying methods in natural language processing on electronic health records (EHR) data is a growing field. Existing corpus and annotation focus on modeling textual features and relation prediction. However, there is a paucity of annotated corpus built to model clinical diagnostic thinking, a process involving text understanding, domain knowledge abstraction and reasoning. This work introduces a hierarchical annotation schema with three stages to address clinical text understanding, clinical reasoning, and summarization. We created an annotated corpus based on an extensive collection of publicly available daily progress notes, a type of EHR documentation that is collected in time series in a problem-oriented format. The conventional format for a progress note follows a Subjective, Objective, Assessment and Plan heading (SOAP). We also define a new suite of tasks, Progress Note Understanding, with three tasks utilizing the three annotation stages. The novel suite of tasks was designed to train and evaluate future NLP models for clinical text understanding, clinical knowledge representation, inference, and summarization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge