Matthew M. Churpek
MoMA: A Mixture-of-Multimodal-Agents Architecture for Enhancing Clinical Prediction Modelling
Aug 07, 2025Abstract:Multimodal electronic health record (EHR) data provide richer, complementary insights into patient health compared to single-modality data. However, effectively integrating diverse data modalities for clinical prediction modeling remains challenging due to the substantial data requirements. We introduce a novel architecture, Mixture-of-Multimodal-Agents (MoMA), designed to leverage multiple large language model (LLM) agents for clinical prediction tasks using multimodal EHR data. MoMA employs specialized LLM agents ("specialist agents") to convert non-textual modalities, such as medical images and laboratory results, into structured textual summaries. These summaries, together with clinical notes, are combined by another LLM ("aggregator agent") to generate a unified multimodal summary, which is then used by a third LLM ("predictor agent") to produce clinical predictions. Evaluating MoMA on three prediction tasks using real-world datasets with different modality combinations and prediction settings, MoMA outperforms current state-of-the-art methods, highlighting its enhanced accuracy and flexibility across various tasks.
Leveraging A Medical Knowledge Graph into Large Language Models for Diagnosis Prediction
Aug 28, 2023



Abstract:Electronic Health Records (EHRs) and routine documentation practices play a vital role in patients' daily care, providing a holistic record of health, diagnoses, and treatment. However, complex and verbose EHR narratives overload healthcare providers, risking diagnostic inaccuracies. While Large Language Models (LLMs) have showcased their potential in diverse language tasks, their application in the healthcare arena needs to ensure the minimization of diagnostic errors and the prevention of patient harm. In this paper, we outline an innovative approach for augmenting the proficiency of LLMs in the realm of automated diagnosis generation, achieved through the incorporation of a medical knowledge graph (KG) and a novel graph model: Dr.Knows, inspired by the clinical diagnostic reasoning process. We derive the KG from the National Library of Medicine's Unified Medical Language System (UMLS), a robust repository of biomedical knowledge. Our method negates the need for pre-training and instead leverages the KG as an auxiliary instrument aiding in the interpretation and summarization of complex medical concepts. Using real-world hospital datasets, our experimental results demonstrate that the proposed approach of combining LLMs with KG has the potential to improve the accuracy of automated diagnosis generation. More importantly, our approach offers an explainable diagnostic pathway, edging us closer to the realization of AI-augmented diagnostic decision support systems.
Multi-Task Training with In-Domain Language Models for Diagnostic Reasoning
Jun 13, 2023Abstract:Generative artificial intelligence (AI) is a promising direction for augmenting clinical diagnostic decision support and reducing diagnostic errors, a leading contributor to medical errors. To further the development of clinical AI systems, the Diagnostic Reasoning Benchmark (DR.BENCH) was introduced as a comprehensive generative AI framework, comprised of six tasks representing key components in clinical reasoning. We present a comparative analysis of in-domain versus out-of-domain language models as well as multi-task versus single task training with a focus on the problem summarization task in DR.BENCH (Gao et al., 2023). We demonstrate that a multi-task, clinically trained language model outperforms its general domain counterpart by a large margin, establishing a new state-of-the-art performance, with a ROUGE-L score of 28.55. This research underscores the value of domain-specific training for optimizing clinical diagnostic reasoning tasks.
Overview of the Problem List Summarization (ProbSum) 2023 Shared Task on Summarizing Patients' Active Diagnoses and Problems from Electronic Health Record Progress Notes
Jun 08, 2023Abstract:The BioNLP Workshop 2023 initiated the launch of a shared task on Problem List Summarization (ProbSum) in January 2023. The aim of this shared task is to attract future research efforts in building NLP models for real-world diagnostic decision support applications, where a system generating relevant and accurate diagnoses will augment the healthcare providers decision-making process and improve the quality of care for patients. The goal for participants is to develop models that generated a list of diagnoses and problems using input from the daily care notes collected from the hospitalization of critically ill patients. Eight teams submitted their final systems to the shared task leaderboard. In this paper, we describe the tasks, datasets, evaluation metrics, and baseline systems. Additionally, the techniques and results of the evaluation of the different approaches tried by the participating teams are summarized.
Summarizing Patients Problems from Hospital Progress Notes Using Pre-trained Sequence-to-Sequence Models
Aug 17, 2022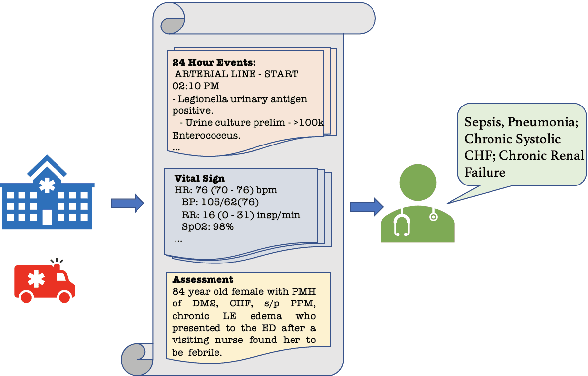
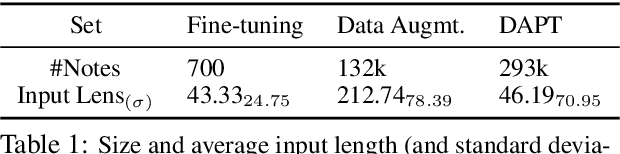
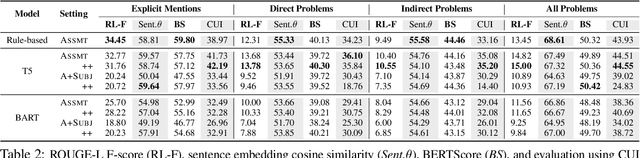
Abstract:Automatically summarizing patients' main problems from daily progress notes using natural language processing methods helps to battle against information and cognitive overload in hospital settings and potentially assists providers with computerized diagnostic decision support. Problem list summarization requires a model to understand, abstract, and generate clinical documentation. In this work, we propose a new NLP task that aims to generate a list of problems in a patient's daily care plan using input from the provider's progress notes during hospitalization. We investigate the performance of T5 and BART, two state-of-the-art seq2seq transformer architectures, in solving this problem. We provide a corpus built on top of progress notes from publicly available electronic health record progress notes in the Medical Information Mart for Intensive Care (MIMIC)-III. T5 and BART are trained on general domain text, and we experiment with a data augmentation method and a domain adaptation pre-training method to increase exposure to medical vocabulary and knowledge. Evaluation methods include ROUGE, BERTScore, cosine similarity on sentence embedding, and F-score on medical concepts. Results show that T5 with domain adaptive pre-training achieves significant performance gains compared to a rule-based system and general domain pre-trained language models, indicating a promising direction for tackling the problem summarization task.
Hierarchical Annotation for Building A Suite of Clinical Natural Language Processing Tasks: Progress Note Understanding
Apr 06, 2022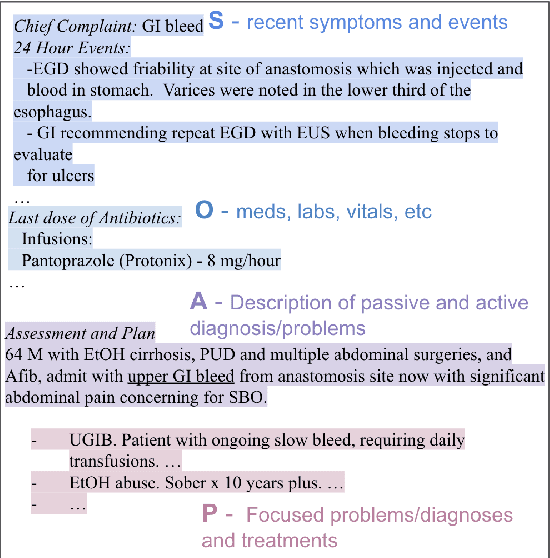
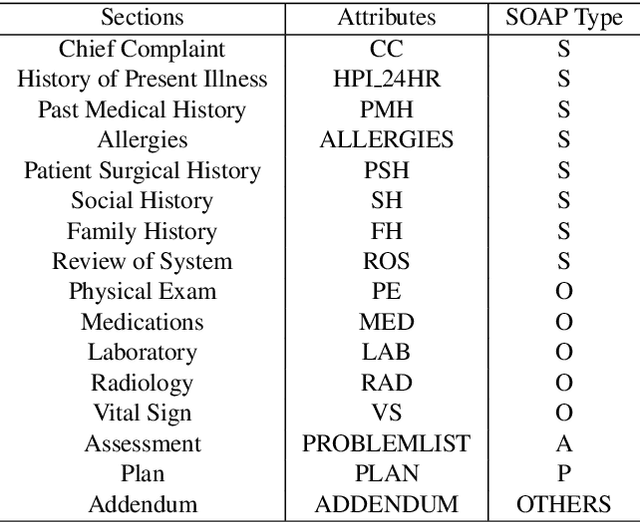
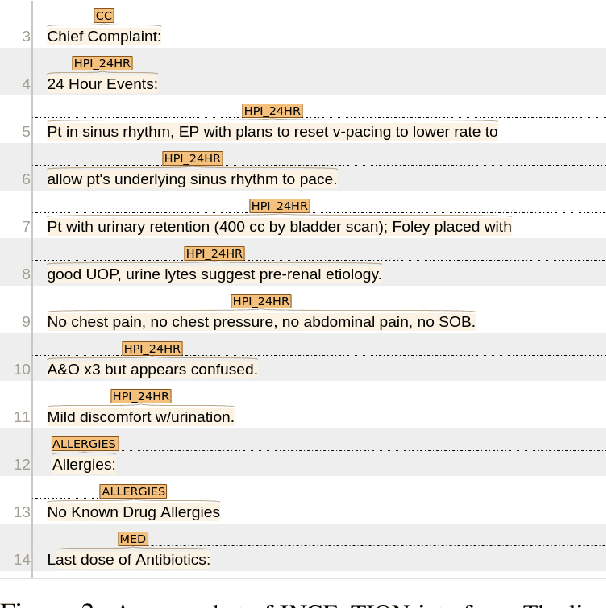
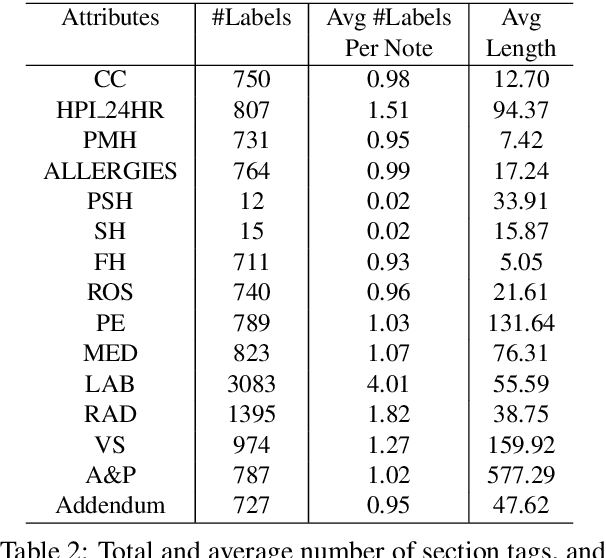
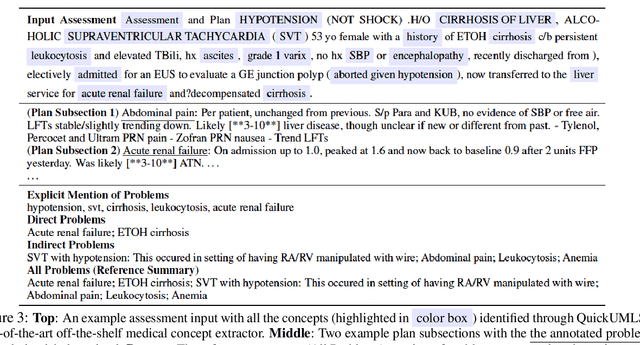
Abstract:Applying methods in natural language processing on electronic health records (EHR) data is a growing field. Existing corpus and annotation focus on modeling textual features and relation prediction. However, there is a paucity of annotated corpus built to model clinical diagnostic thinking, a process involving text understanding, domain knowledge abstraction and reasoning. This work introduces a hierarchical annotation schema with three stages to address clinical text understanding, clinical reasoning, and summarization. We created an annotated corpus based on an extensive collection of publicly available daily progress notes, a type of EHR documentation that is collected in time series in a problem-oriented format. The conventional format for a progress note follows a Subjective, Objective, Assessment and Plan heading (SOAP). We also define a new suite of tasks, Progress Note Understanding, with three tasks utilizing the three annotation stages. The novel suite of tasks was designed to train and evaluate future NLP models for clinical text understanding, clinical knowledge representation, inference, and summarization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge