Thomas Wollmann
Float8@2bits: Entropy Coding Enables Data-Free Model Compression
Jan 30, 2026Abstract:Post-training compression is currently divided into two contrasting regimes. On the one hand, fast, data-free, and model-agnostic methods (e.g., NF4 or HQQ) offer maximum accessibility but suffer from functional collapse at extreme bit-rates below 4 bits. On the other hand, techniques leveraging calibration data or extensive recovery training achieve superior fidelity but impose high computational constraints and face uncertain robustness under data distribution shifts. We introduce EntQuant, the first framework to unite the advantages of these distinct paradigms. By matching the performance of data-dependent methods with the speed and universality of data-free techniques, EntQuant enables practical utility in the extreme compression regime. Our method decouples numerical precision from storage cost via entropy coding, compressing a 70B parameter model in less than 30 minutes. We demonstrate that EntQuant does not only achieve state-of-the-art results on standard evaluation sets and models, but also retains functional performance on more complex benchmarks with instruction-tuned models, all at modest inference overhead.
Choose Your Model Size: Any Compression by a Single Gradient Descent
Feb 03, 2025



Abstract:The adoption of Foundation Models in resource-constrained environments remains challenging due to their large size and inference costs. A promising way to overcome these limitations is post-training compression, which aims to balance reduced model size against performance degradation. This work presents Any Compression via Iterative Pruning (ACIP), a novel algorithmic approach to determine a compression-performance trade-off from a single stochastic gradient descent run. To ensure parameter efficiency, we use an SVD-reparametrization of linear layers and iteratively prune their singular values with a sparsity-inducing penalty. The resulting pruning order gives rise to a global parameter ranking that allows us to materialize models of any target size. Importantly, the compressed models exhibit strong predictive downstream performance without the need for costly fine-tuning. We evaluate ACIP on a large selection of open-weight LLMs and tasks, and demonstrate state-of-the-art results compared to existing factorisation-based compression methods. We also show that ACIP seamlessly complements common quantization-based compression techniques.
MEAL: Manifold Embedding-based Active Learning
Jul 20, 2021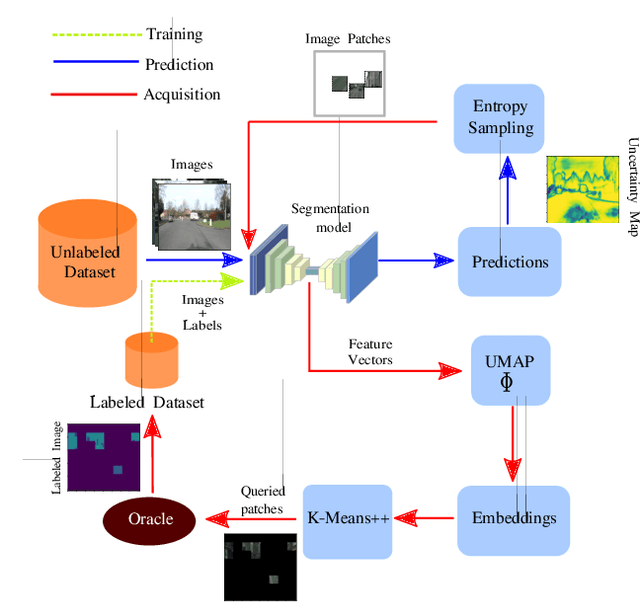
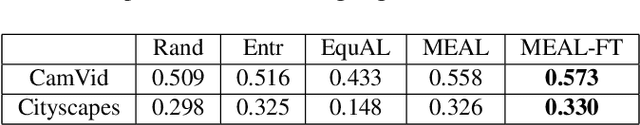
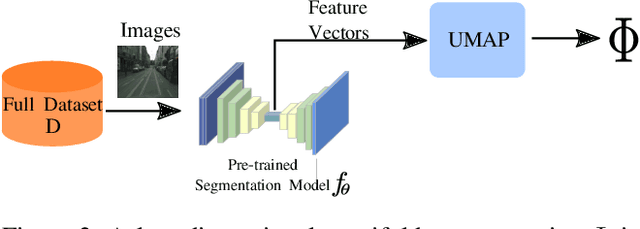
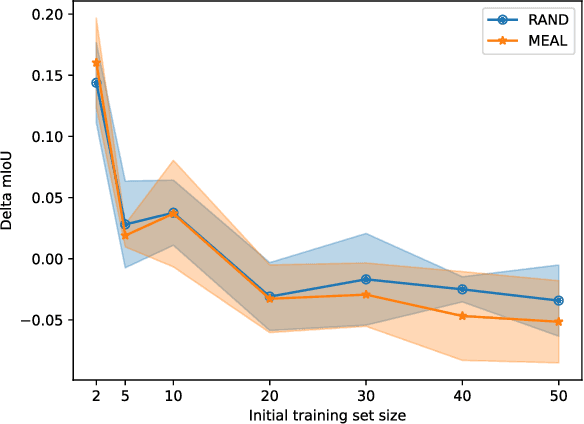
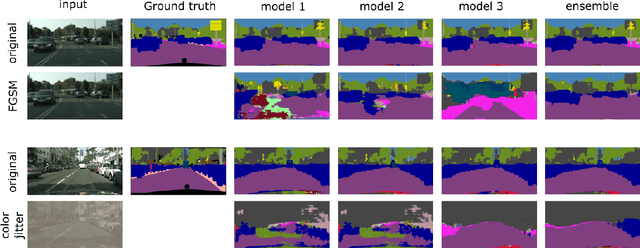
Abstract:Image segmentation is a common and challenging task in autonomous driving. Availability of sufficient pixel-level annotations for the training data is a hurdle. Active learning helps learning from small amounts of data by suggesting the most promising samples for labeling. In this work, we propose a new pool-based method for active learning, which proposes promising patches extracted from full image, in each acquisition step. The problem is framed in an exploration-exploitation framework by combining an embedding based on Uniform Manifold Approximation to model representativeness with entropy as uncertainty measure to model informativeness. We applied our proposed method to the autonomous driving datasets CamVid and Cityscapes and performed a quantitative comparison with state-of-the-art baselines. We find that our active learning method achieves better performance compared to previous methods.
DAAIN: Detection of Anomalous and Adversarial Input using Normalizing Flows
May 30, 2021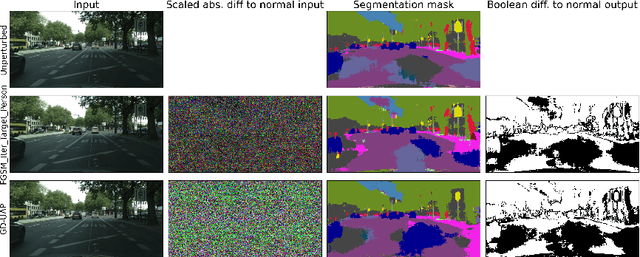
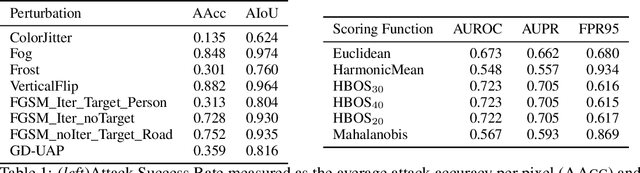
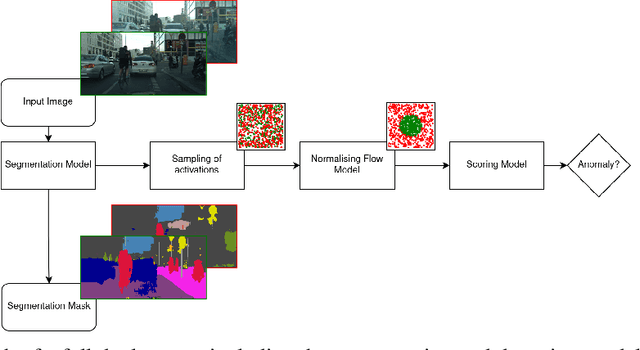
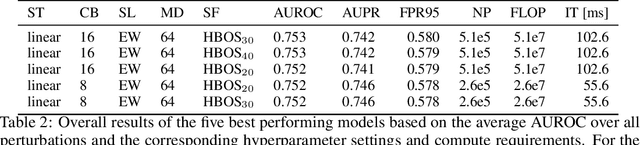
Abstract:Despite much recent work, detecting out-of-distribution (OOD) inputs and adversarial attacks (AA) for computer vision models remains a challenge. In this work, we introduce a novel technique, DAAIN, to detect OOD inputs and AA for image segmentation in a unified setting. Our approach monitors the inner workings of a neural network and learns a density estimator of the activation distribution. We equip the density estimator with a classification head to discriminate between regular and anomalous inputs. To deal with the high-dimensional activation-space of typical segmentation networks, we subsample them to obtain a homogeneous spatial and layer-wise coverage. The subsampling pattern is chosen once per monitored model and kept fixed for all inputs. Since the attacker has access to neither the detection model nor the sampling key, it becomes harder for them to attack the segmentation network, as the attack cannot be backpropagated through the detector. We demonstrate the effectiveness of our approach using an ESPNet trained on the Cityscapes dataset as segmentation model, an affine Normalizing Flow as density estimator and use blue noise to ensure homogeneous sampling. Our model can be trained on a single GPU making it compute efficient and deployable without requiring specialized accelerators.
Chameleon: A Semi-AutoML framework targeting quick and scalable development and deployment of production-ready ML systems for SMEs
May 08, 2021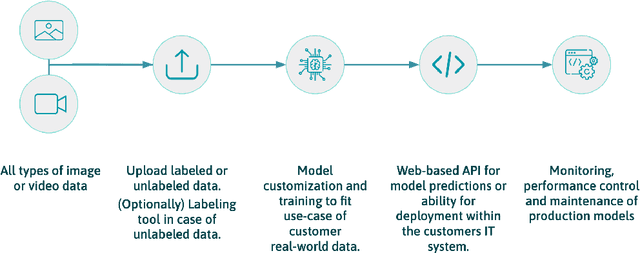

Abstract:Developing, scaling, and deploying modern Machine Learning solutions remains challenging for small- and middle-sized enterprises (SMEs). This is due to a high entry barrier of building and maintaining a dedicated IT team as well as the difficulties of real-world data (RWD) compared to standard benchmark data. To address this challenge, we discuss the implementation and concepts of Chameleon, a semi-AutoML framework. The goal of Chameleon is fast and scalable development and deployment of production-ready machine learning systems into the workflow of SMEs. We first discuss the RWD challenges faced by SMEs. After, we outline the central part of the framework which is a model and loss-function zoo with RWD-relevant defaults. Subsequently, we present how one can use a templatable framework in order to automate the experiment iteration cycle, as well as close the gap between development and deployment. Finally, we touch on our testing framework component allowing us to investigate common model failure modes and support best practices of model deployment governance.
Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge
Jul 22, 2018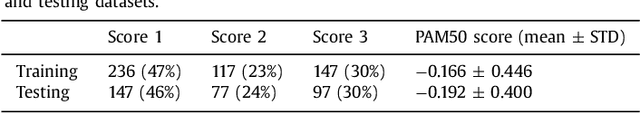
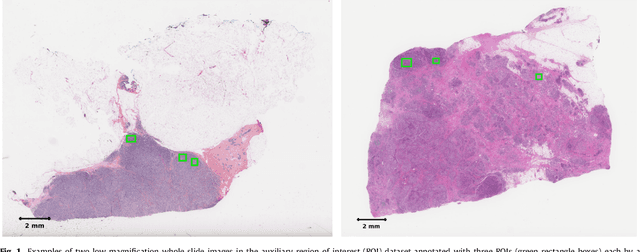
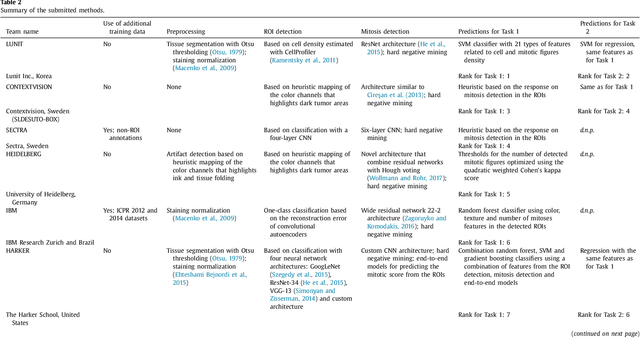
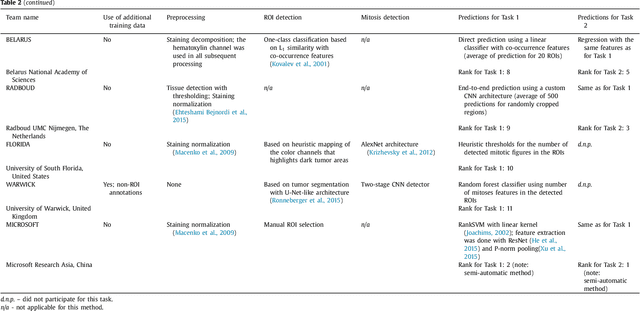
Abstract:Tumor proliferation is an important biomarker indicative of the prognosis of breast cancer patients. Assessment of tumor proliferation in a clinical setting is highly subjective and labor-intensive task. Previous efforts to automate tumor proliferation assessment by image analysis only focused on mitosis detection in predefined tumor regions. However, in a real-world scenario, automatic mitosis detection should be performed in whole-slide images (WSIs) and an automatic method should be able to produce a tumor proliferation score given a WSI as input. To address this, we organized the TUmor Proliferation Assessment Challenge 2016 (TUPAC16) on prediction of tumor proliferation scores from WSIs. The challenge dataset consisted of 500 training and 321 testing breast cancer histopathology WSIs. In order to ensure fair and independent evaluation, only the ground truth for the training dataset was provided to the challenge participants. The first task of the challenge was to predict mitotic scores, i.e., to reproduce the manual method of assessing tumor proliferation by a pathologist. The second task was to predict the gene expression based PAM50 proliferation scores from the WSI. The best performing automatic method for the first task achieved a quadratic-weighted Cohen's kappa score of $\kappa$ = 0.567, 95% CI [0.464, 0.671] between the predicted scores and the ground truth. For the second task, the predictions of the top method had a Spearman's correlation coefficient of r = 0.617, 95% CI [0.581 0.651] with the ground truth. This was the first study that investigated tumor proliferation assessment from WSIs. The achieved results are promising given the difficulty of the tasks and weakly-labelled nature of the ground truth. However, further research is needed to improve the practical utility of image analysis methods for this task.
Automatic breast cancer grading in lymph nodes using a deep neural network
Jul 24, 2017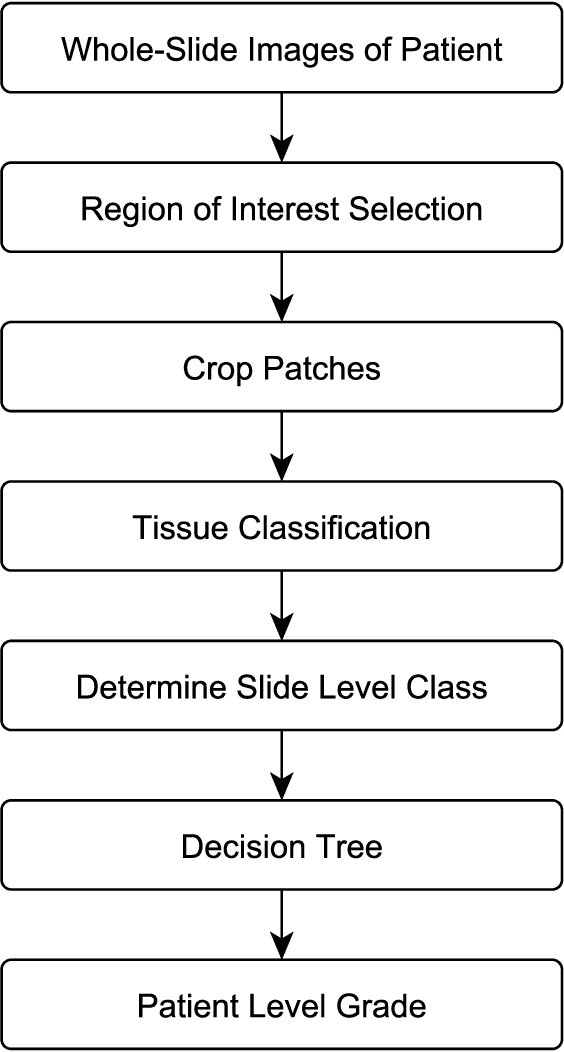

Abstract:The progression of breast cancer can be quantified in lymph node whole-slide images (WSIs). We describe a novel method for effectively performing classification of whole-slide images and patient level breast cancer grading. Our method utilises a deep neural network. The method performs classification on small patches and uses model averaging for boosting. In the first step, region of interest patches are determined and cropped automatically by color thresholding and then classified by the deep neural network. The classification results are used to determine a slide level class and for further aggregation to predict a patient level grade. Fast processing speed of our method enables high throughput image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge