Tarcisio Mendes de Farias
SIB Swiss Institute of Bioinformatics, Switzerland
SPARQL-LLM: Real-Time SPARQL Query Generation from Natural Language Questions
Dec 16, 2025Abstract:The advent of large language models is contributing to the emergence of novel approaches that promise to better tackle the challenge of generating structured queries, such as SPARQL queries, from natural language. However, these new approaches mostly focus on response accuracy over a single source while ignoring other evaluation criteria, such as federated query capability over distributed data stores, as well as runtime and cost to generate SPARQL queries. Consequently, they are often not production-ready or easy to deploy over (potentially federated) knowledge graphs with good accuracy. To mitigate these issues, in this paper, we extend our previous work and describe and systematically evaluate SPARQL-LLM, an open-source and triplestore-agnostic approach, powered by lightweight metadata, that generates SPARQL queries from natural language text. First, we describe its architecture, which consists of dedicated components for metadata indexing, prompt building, and query generation and execution. Then, we evaluate it based on a state-of-the-art challenge with multilingual questions, and a collection of questions from three of the most prevalent knowledge graphs within the field of bioinformatics. Our results demonstrate a substantial increase of 24% in the F1 Score on the state-of-the-art challenge, adaptability to high-resource languages such as English and Spanish, as well as ability to form complex and federated bioinformatics queries. Furthermore, we show that SPARQL-LLM is up to 36x faster than other systems participating in the challenge, while costing a maximum of $0.01 per question, making it suitable for real-time, low-cost text-to-SPARQL applications. One such application deployed over real-world decentralized knowledge graphs can be found at https://www.expasy.org/chat.
LLM-based SPARQL Query Generation from Natural Language over Federated Knowledge Graphs
Oct 10, 2024Abstract:We introduce a Retrieval-Augmented Generation (RAG) system for translating user questions into accurate federated SPARQL queries over bioinformatics knowledge graphs (KGs) leveraging Large Language Models (LLMs). To enhance accuracy and reduce hallucinations in query generation, our system utilises metadata from the KGs, including query examples and schema information, and incorporates a validation step to correct generated queries. The system is available online at chat.expasy.org.
A large collection of bioinformatics question-query pairs over federated knowledge graphs: methodology and applications
Oct 08, 2024



Abstract:Background. In the last decades, several life science resources have structured data using the same framework and made these accessible using the same query language to facilitate interoperability. Knowledge graphs have seen increased adoption in bioinformatics due to their advantages for representing data in a generic graph format. For example, yummydata.org catalogs more than 60 knowledge graphs accessible through SPARQL, a technical query language. Although SPARQL allows powerful, expressive queries, even across physically distributed knowledge graphs, formulating such queries is a challenge for most users. Therefore, to guide users in retrieving the relevant data, many of these resources provide representative examples. These examples can also be an important source of information for machine learning, if a sufficiently large number of examples are provided and published in a common, machine-readable and standardized format across different resources. Findings. We introduce a large collection of human-written natural language questions and their corresponding SPARQL queries over federated bioinformatics knowledge graphs (KGs) collected for several years across different research groups at the SIB Swiss Institute of Bioinformatics. The collection comprises more than 1000 example questions and queries, including 65 federated queries. We propose a methodology to uniformly represent the examples with minimal metadata, based on existing standards. Furthermore, we introduce an extensive set of open-source applications, including query graph visualizations and smart query editors, easily reusable by KG maintainers who adopt the proposed methodology. Conclusions. We encourage the community to adopt and extend the proposed methodology, towards richer KG metadata and improved Semantic Web services.
SPARQL Generation: an analysis on fine-tuning OpenLLaMA for Question Answering over a Life Science Knowledge Graph
Feb 07, 2024


Abstract:The recent success of Large Language Models (LLM) in a wide range of Natural Language Processing applications opens the path towards novel Question Answering Systems over Knowledge Graphs leveraging LLMs. However, one of the main obstacles preventing their implementation is the scarcity of training data for the task of translating questions into corresponding SPARQL queries, particularly in the case of domain-specific KGs. To overcome this challenge, in this study, we evaluate several strategies for fine-tuning the OpenLlama LLM for question answering over life science knowledge graphs. In particular, we propose an end-to-end data augmentation approach for extending a set of existing queries over a given knowledge graph towards a larger dataset of semantically enriched question-to-SPARQL query pairs, enabling fine-tuning even for datasets where these pairs are scarce. In this context, we also investigate the role of semantic "clues" in the queries, such as meaningful variable names and inline comments. Finally, we evaluate our approach over the real-world Bgee gene expression knowledge graph and we show that semantic clues can improve model performance by up to 33% compared to a baseline with random variable names and no comments included.
On the Potential of Artificial Intelligence Chatbots for Data Exploration of Federated Bioinformatics Knowledge Graphs
Apr 20, 2023Abstract:In this paper, we present work in progress on the role of artificial intelligence (AI) chatbots, such as ChatGPT, in facilitating data access to federated knowledge graphs. In particular, we provide examples from the field of bioinformatics, to illustrate the potential use of Conversational AI to describe datasets, as well as generate and explain (federated) queries across datasets for the benefit of domain experts.
INODE: Building an End-to-End Data Exploration System in Practice
Apr 09, 2021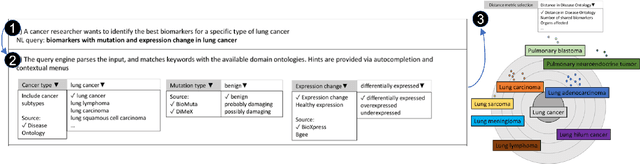
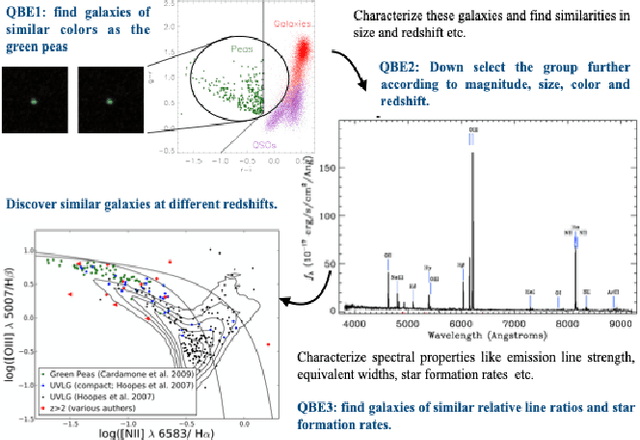
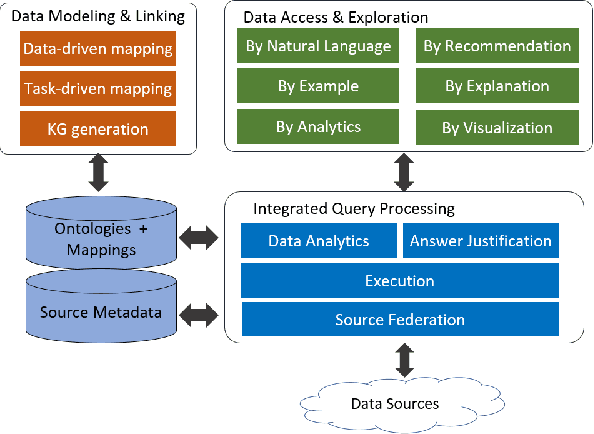
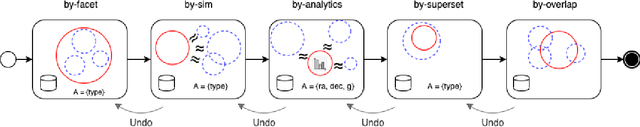
Abstract:A full-fledged data exploration system must combine different access modalities with a powerful concept of guiding the user in the exploration process, by being reactive and anticipative both for data discovery and for data linking. Such systems are a real opportunity for our community to cater to users with different domain and data science expertise. We introduce INODE -- an end-to-end data exploration system -- that leverages, on the one hand, Machine Learning and, on the other hand, semantics for the purpose of Data Management (DM). Our vision is to develop a classic unified, comprehensive platform that provides extensive access to open datasets, and we demonstrate it in three significant use cases in the fields of Cancer Biomarker Reearch, Research and Innovation Policy Making, and Astrophysics. INODE offers sustainable services in (a) data modeling and linking, (b) integrated query processing using natural language, (c) guidance, and (d) data exploration through visualization, thus facilitating the user in discovering new insights. We demonstrate that our system is uniquely accessible to a wide range of users from larger scientific communities to the public. Finally, we briefly illustrate how this work paves the way for new research opportunities in DM.
IfcWoD, Semantically Adapting IFC Model Relations into OWL Properties
Nov 12, 2015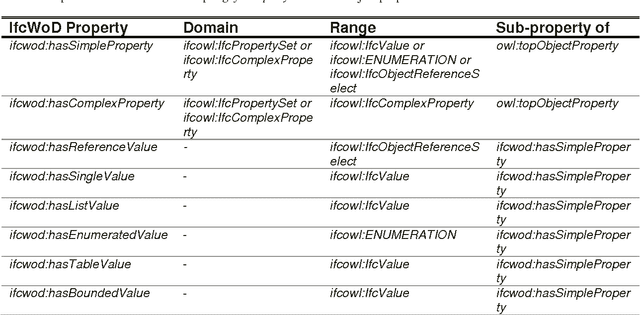
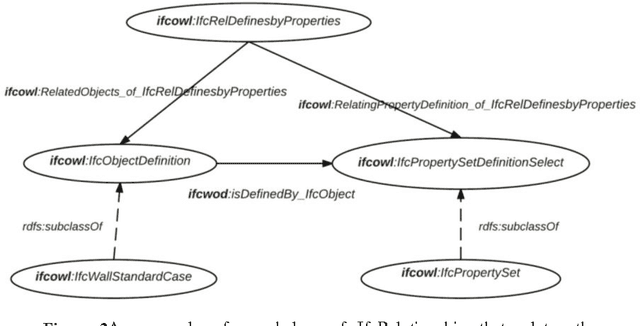
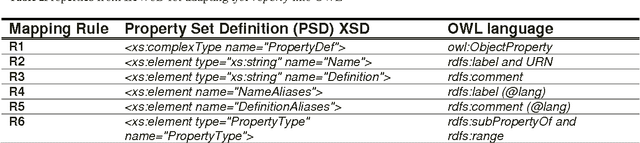

Abstract:In the context of Building Information Modelling, ontologies have been identified as interesting in achieving information interoperability. Regarding the construction and facility management domains, several IFC (Industry Foundation Classes) based ontologies have been developed, such as IfcOWL. In the context of ontology modelling, the constraint of optimizing the size of IFC STEP-based files can be leveraged. In this paper, we propose an adaptation of the IFC model into OWL which leverages from all modelling constraints required by the object-oriented structure of IFC schema. Therefore, we do not only present a syntactic but also a semantic adaptation of the IFC model. Our model takes into consideration the meaning of entities, relationships, properties and attributes defined by the IFC standard. Our approach presents several advantages compared to other initiatives such as the optimization of query execution time. Every advantage is defended by means of practical examples and benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge