Stefano Nichele
A Unified Substrate for Body-Brain Co-evolution
Mar 22, 2022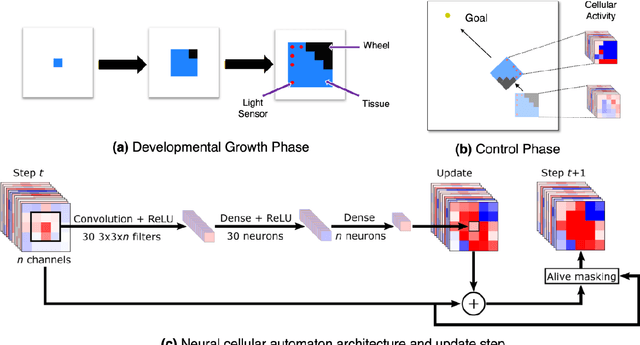
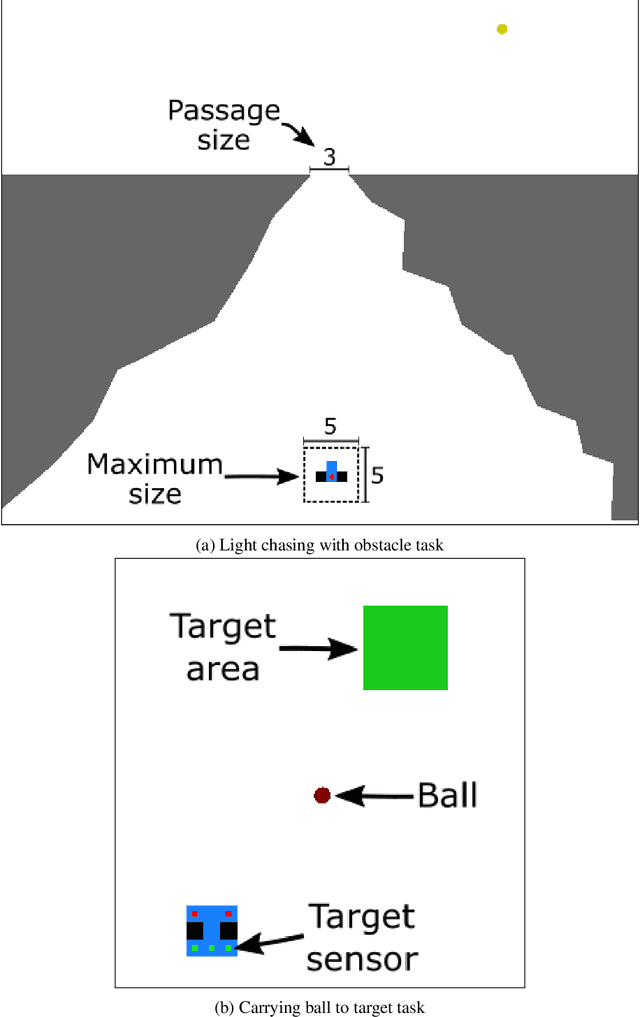
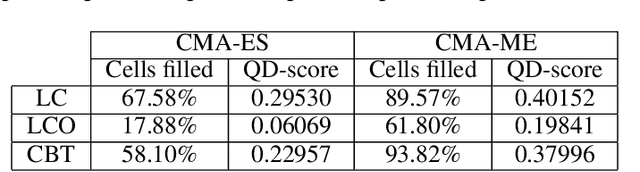
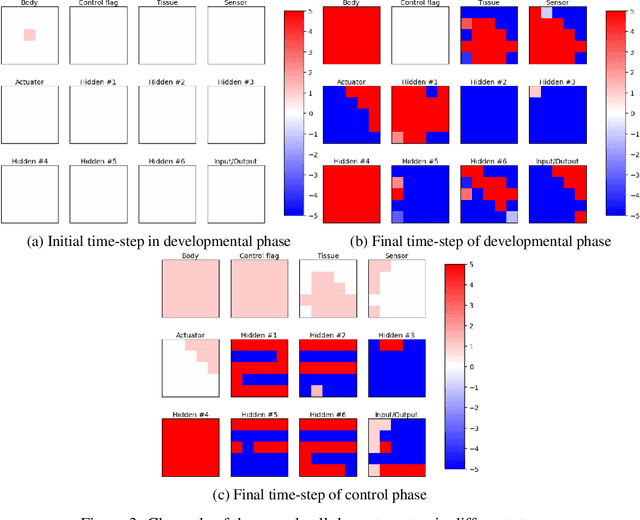
Abstract:A successful development of a complex multicellular organism took millions of years of evolution. The genome of such a multicellular organism guides the development of its body from a single cell, including its control system. Our goal is to imitate this natural process using a single neural cellular automaton (NCA) as a genome for modular robotic agents. In the introduced approach, called Neural Cellular Robot Substrate (NCRS), a single NCA guides the growth of a robot and the cellular activity which controls the robot during deployment. We also introduce three benchmark environments, which test the ability of the approach to grow different robot morphologies. We evolve the NCRS with covariance matrix adaptation evolution strategy (CMA-ES), and covariance matrix adaptation MAP-Elites (CMA-ME) for quality diversity and observe that CMA-ME generates more diverse robot morphologies with higher fitness scores. While the NCRS is able to solve the easier tasks in the benchmark, the success rate reduces when the difficulty of the task increases. We discuss directions for future work that may facilitate the use of the NCRS approach for more complex domains.
AI-based artistic representation of emotions from EEG signals: a discussion on fairness, inclusion, and aesthetics
Feb 07, 2022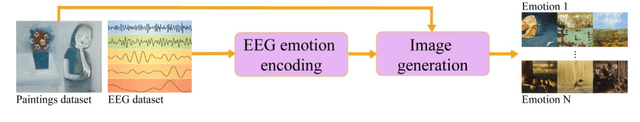


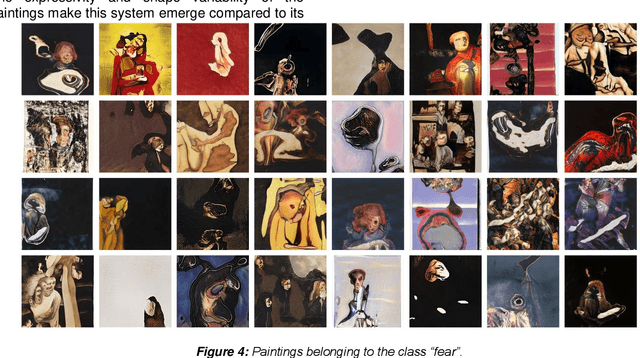
Abstract:While Artificial Intelligence (AI) technologies are being progressively developed, artists and researchers are investigating their role in artistic practices. In this work, we present an AI-based Brain-Computer Interface (BCI) in which humans and machines interact to express feelings artistically. This system and its production of images give opportunities to reflect on the complexities and range of human emotions and their expressions. In this discussion, we seek to understand the dynamics of this interaction to reach better co-existence in fairness, inclusion, and aesthetics.
Evolving spiking neuron cellular automata and networks to emulate in vitro neuronal activity
Oct 15, 2021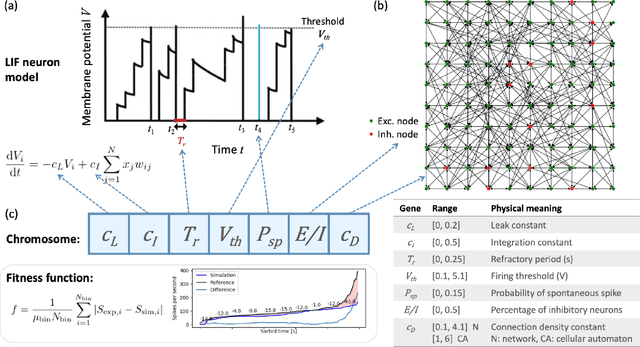
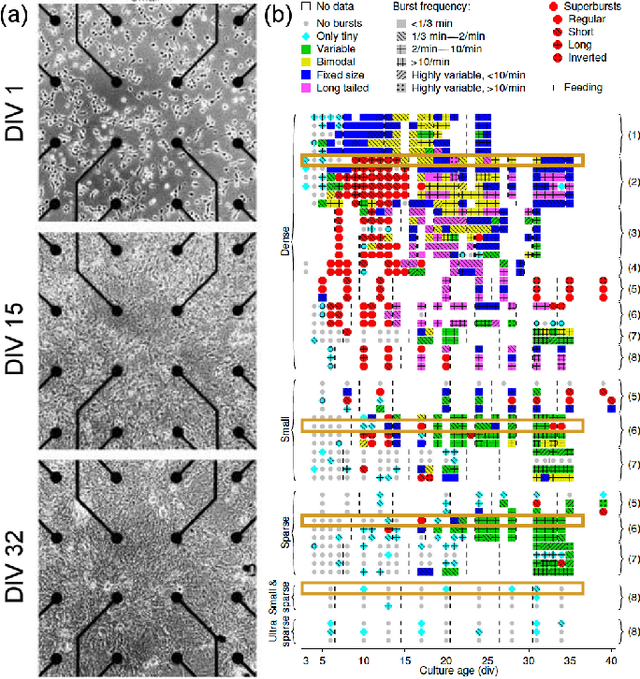
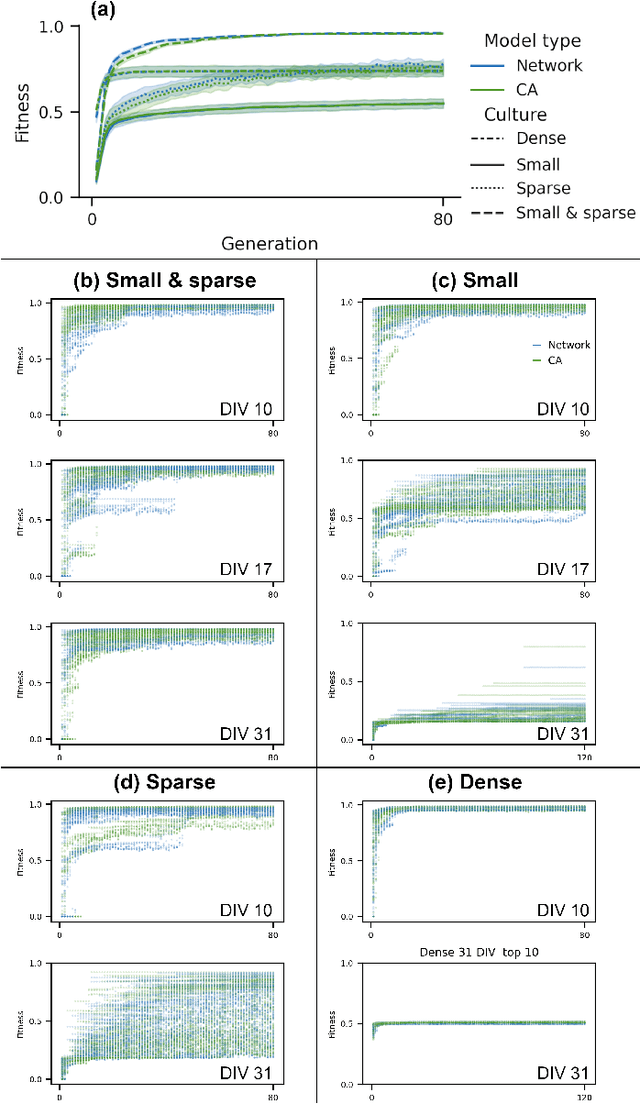
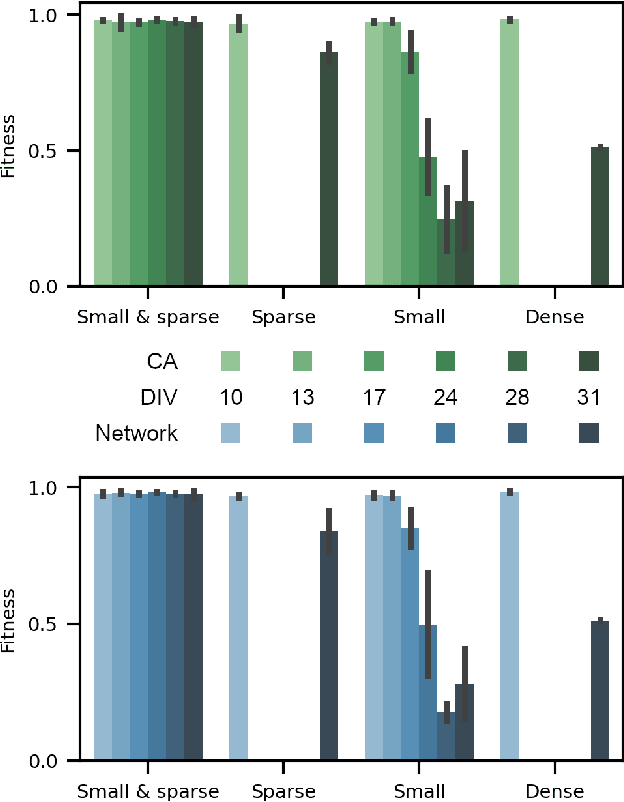
Abstract:Neuro-inspired models and systems have great potential for applications in unconventional computing. Often, the mechanisms of biological neurons are modeled or mimicked in simulated or physical systems in an attempt to harness some of the computational power of the brain. However, the biological mechanisms at play in neural systems are complicated and challenging to capture and engineer; thus, it can be simpler to turn to a data-driven approach to transfer features of neural behavior to artificial substrates. In the present study, we used an evolutionary algorithm (EA) to produce spiking neural systems that emulate the patterns of behavior of biological neurons in vitro. The aim of this approach was to develop a method of producing models capable of exhibiting complex behavior that may be suitable for use as computational substrates. Our models were able to produce a level of network-wide synchrony and showed a range of behaviors depending on the target data used for their evolution, which was from a range of neuronal culture densities and maturities. The genomes of the top-performing models indicate the excitability and density of connections in the model play an important role in determining the complexity of the produced activity.
Towards self-organized control: Using neural cellular automata to robustly control a cart-pole agent
Jul 12, 2021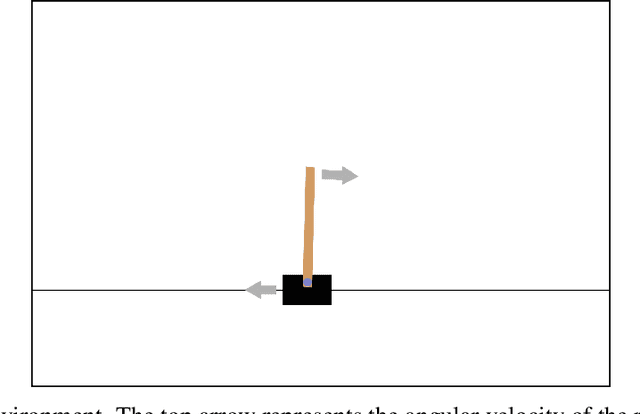

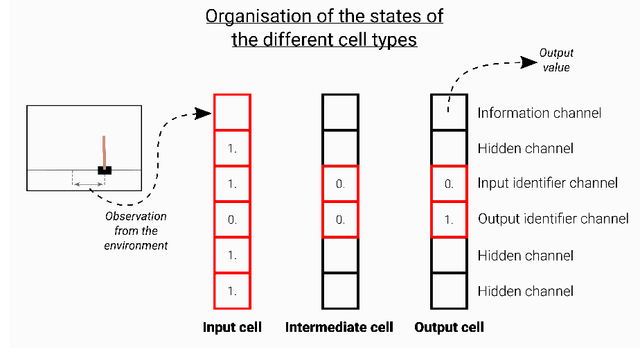

Abstract:Neural cellular automata (Neural CA) are a recent framework used to model biological phenomena emerging from multicellular organisms. In these systems, artificial neural networks are used as update rules for cellular automata. Neural CA are end-to-end differentiable systems where the parameters of the neural network can be learned to achieve a particular task. In this work, we used neural CA to control a cart-pole agent. The observations of the environment are transmitted in input cells, while the values of output cells are used as a readout of the system. We trained the model using deep-Q learning, where the states of the output cells were used as the Q-value estimates to be optimized. We found that the computing abilities of the cellular automata were maintained over several hundreds of thousands of iterations, producing an emergent stable behavior in the environment it controls for thousands of steps. Moreover, the system demonstrated life-like phenomena such as a developmental phase, regeneration after damage, stability despite a noisy environment, and robustness to unseen disruption such as input deletion.
On Artificial Life and Emergent Computation in Physical Substrates
Sep 09, 2020
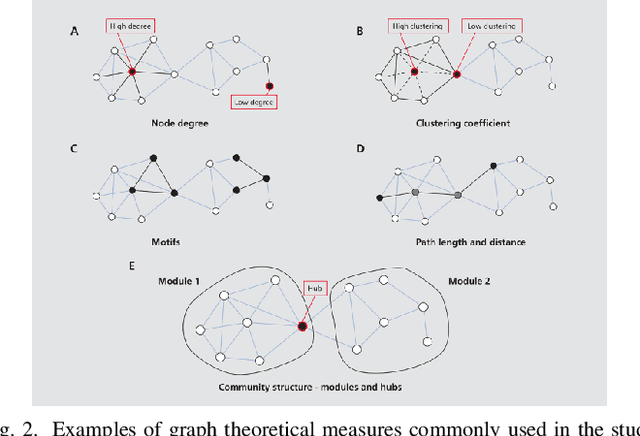
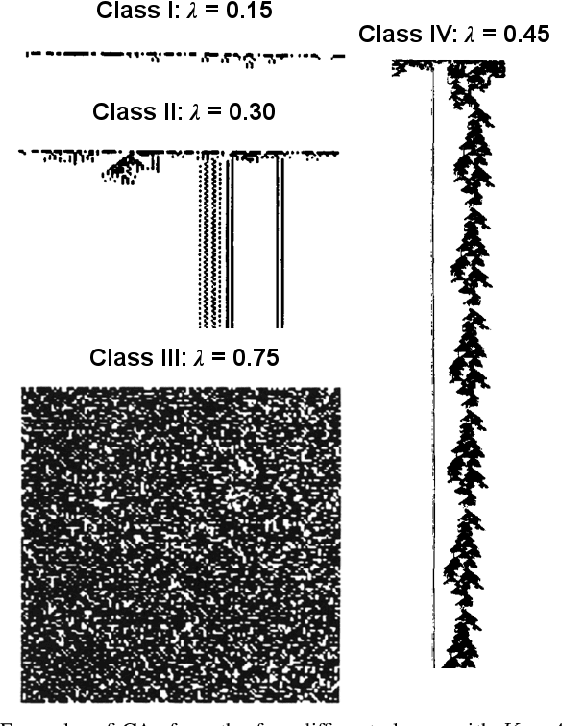
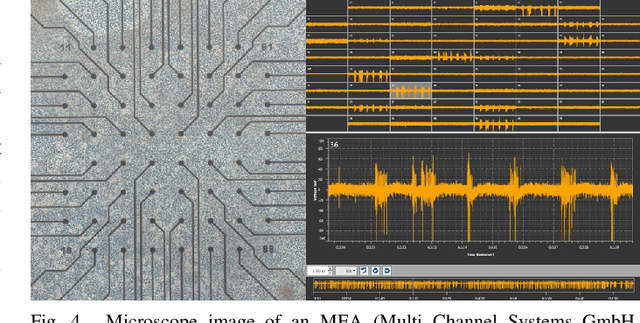
Abstract:In living systems, we often see the emergence of the ingredients necessary for computation -- the capacity for information transmission, storage, and modification -- begging the question of how we may exploit or imitate such biological systems in unconventional computing applications. What can we gain from artificial life in the advancement of computing technology? Artificial life provides us with powerful tools for understanding the dynamic behavior of biological systems and capturing this behavior in manmade substrates. With this approach, we can move towards a new computing paradigm concerned with harnessing emergent computation in physical substrates not governed by the constraints of Moore's law and ultimately realize massively parallel and distributed computing technology. In this paper, we argue that the lens of artificial life offers valuable perspectives for the advancement of high-performance computing technology. We first present a brief foundational background on artificial life and some relevant tools that may be applicable to unconventional computing. Two specific substrates are then discussed in detail: biological neurons and ensembles of nanomagnets. These substrates are the focus of the authors' ongoing work, and they are illustrative of the two sides of the approach outlined here -- the close study of living systems and the construction of artificial systems to produce life-like behaviors. We conclude with a philosophical discussion on what we can learn from approaching computation with the curiosity inherent to the study of artificial life. The main contribution of this paper is to present the great potential of using artificial life methodologies to uncover and harness the inherent computational power of physical substrates toward applications in unconventional high-performance computing.
A deep learning based tool for automatic brain extraction from functional magnetic resonance images in rodents
Dec 06, 2019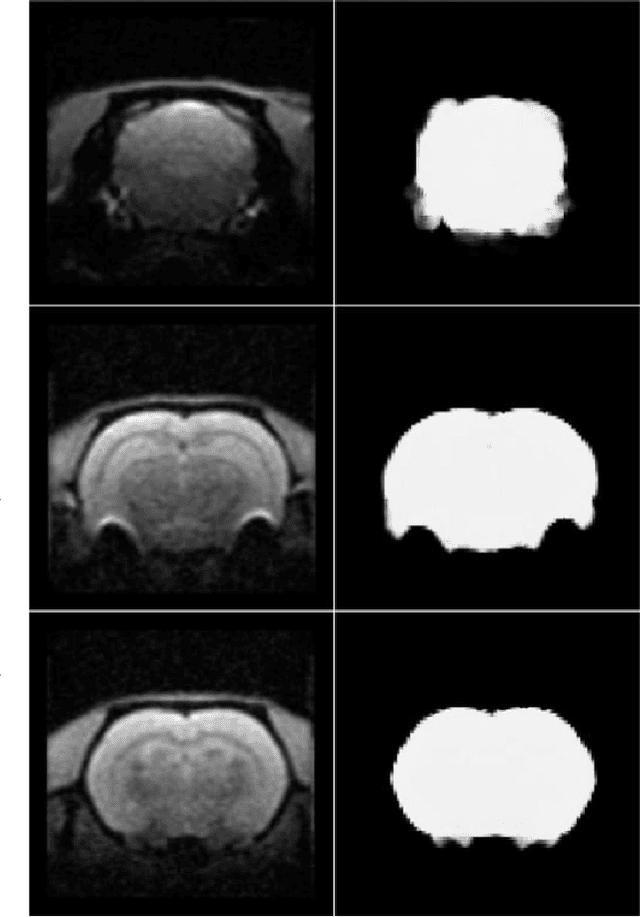
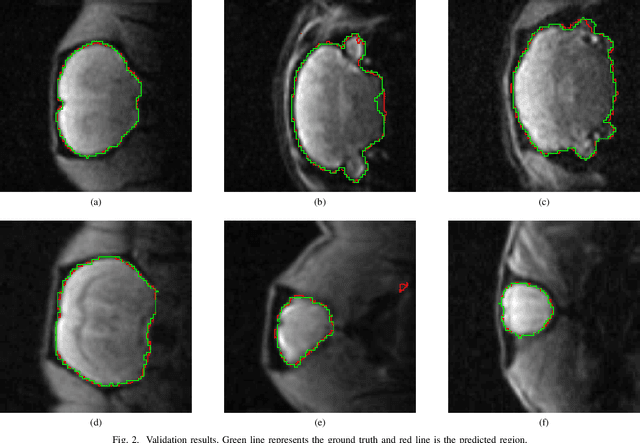
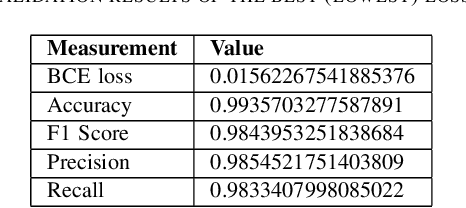
Abstract:Removing skull artifacts from functional magnetic images (fMRI) is a well understood and frequently encountered problem. Because the fMRI field has grown mostly due to human studies, many new tools were developed to handle human data. Nonetheless, these tools are not equally useful to handle the data derived from animal studies, especially from rodents. This represents a major problem to the field because rodent studies generate larger datasets from larger populations, which implies that preprocessing these images manually to remove the skull becomes a bottleneck in the data analysis pipeline. In this study, we address this problem by implementing a neural network based method that uses a U-Net architecture to segment the brain area into a mask and removing the skull and other tissues from the image. We demonstrate several strategies to speed up the process of generating the training dataset using watershedding and several strategies for data augmentation that allowed to train faster the U-Net to perform the segmentation. Finally, we deployed the trained network freely available.
A general representation of dynamical systems for reservoir computing
Jul 03, 2019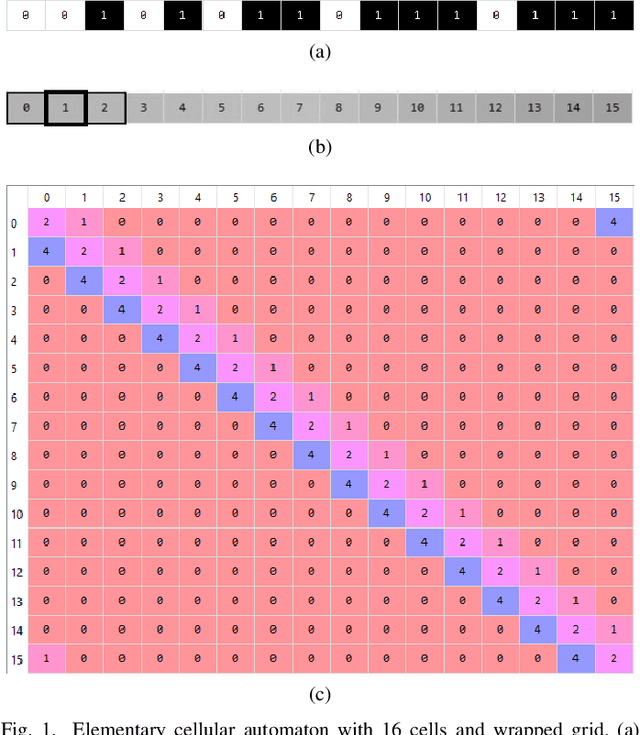
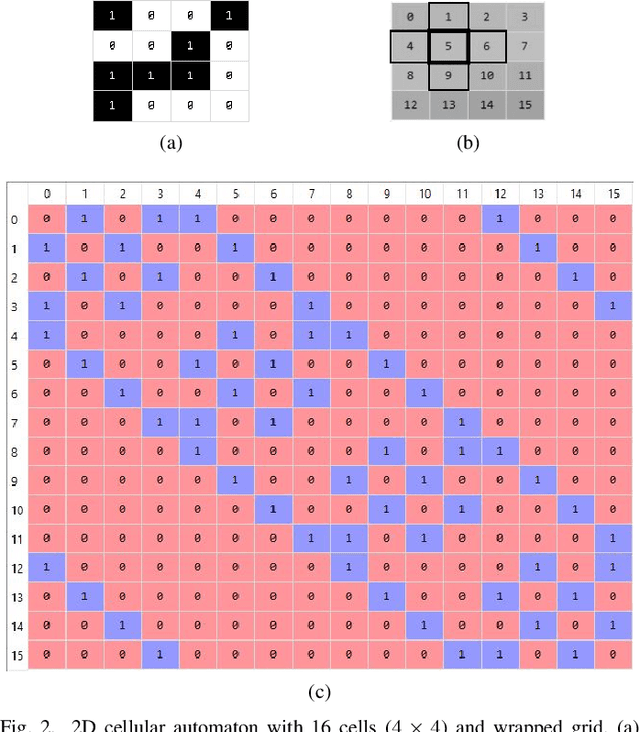
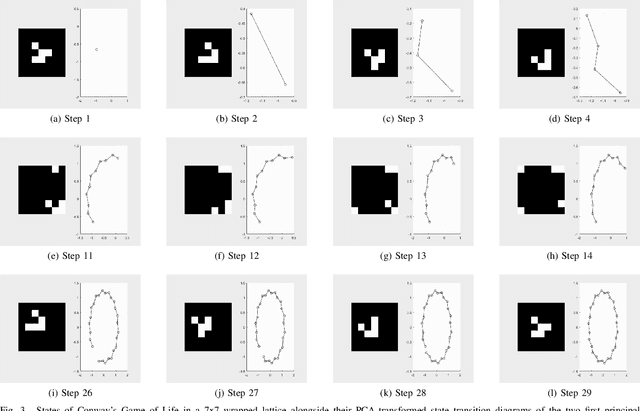
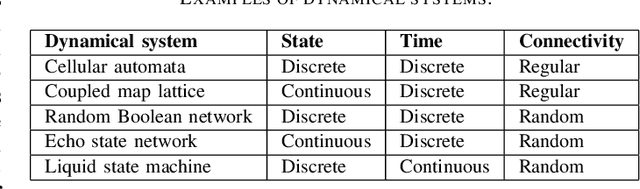
Abstract:Dynamical systems are capable of performing computation in a reservoir computing paradigm. This paper presents a general representation of these systems as an artificial neural network (ANN). Initially, we implement the simplest dynamical system, a cellular automaton. The mathematical fundamentals behind an ANN are maintained, but the weights of the connections and the activation function are adjusted to work as an update rule in the context of cellular automata. The advantages of such implementation are its usage on specialized and optimized deep learning libraries, the capabilities to generalize it to other types of networks and the possibility to evolve cellular automata and other dynamical systems in terms of connectivity, update and learning rules. Our implementation of cellular automata constitutes an initial step towards a general framework for dynamical systems. It aims to evolve such systems to optimize their usage in reservoir computing and to model physical computing substrates.
DeepTEGINN: Deep Learning Based Tools to Extract Graphs from Images of Neural Networks
Jul 01, 2019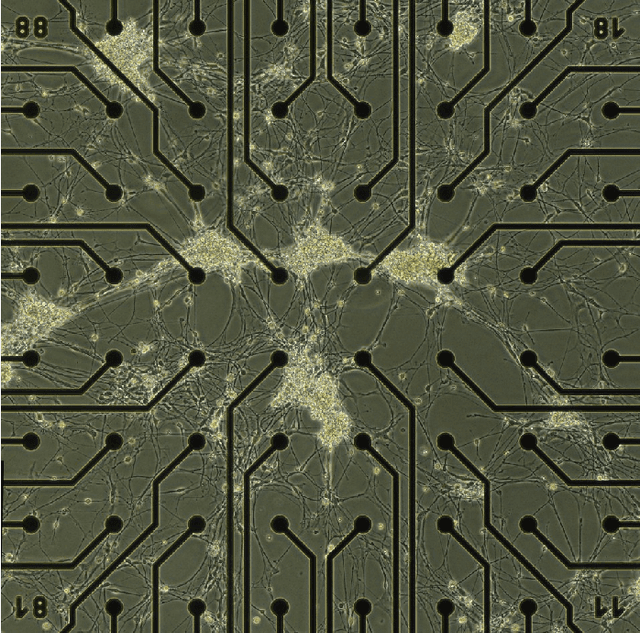

Abstract:In the brain, the structure of a network of neurons defines how these neurons implement the computations that underlie the mind and the behavior of animals and humans. Provided that we can describe the network of neurons as a graph, we can employ methods from graph theory to investigate its structure or use cellular automata to mathematically assess its function. Although, software for the analysis of graphs and cellular automata are widely available. Graph extraction from the image of networks of brain cells remains difficult. Nervous tissue is heterogeneous, and differences in anatomy may reflect relevant differences in function. Here we introduce a deep learning based toolbox to extracts graphs from images of brain tissue. This toolbox provides an easy-to-use framework allowing system neuroscientists to generate graphs based on images of brain tissue by combining methods from image processing, deep learning, and graph theory. The goals are to simplify the training and usage of deep learning methods for computer vision and facilitate its integration into graph extraction pipelines. In this way, the toolbox provides an alternative to the required laborious manual process of tracing, sorting and classifying. We expect to democratize the machine learning methods to a wider community of users beyond the computer vision experts and improve the time-efficiency of graph extraction from large brain image datasets, which may lead to further understanding of the human mind.
Ethics of Artificial Intelligence Demarcations
May 16, 2019
Abstract:In this paper we present a set of key demarcations, particularly important when discussing ethical and societal issues of current AI research and applications. Properly distinguishing issues and concerns related to Artificial General Intelligence and weak AI, between symbolic and connectionist AI, AI methods, data and applications are prerequisites for an informed debate. Such demarcations would not only facilitate much-needed discussions on ethics on current AI technologies and research. In addition sufficiently establishing such demarcations would also enhance knowledge-sharing and support rigor in interdisciplinary research between technical and social sciences.
Evolved Art with Transparent, Overlapping, and Geometric Shapes
May 16, 2019



Abstract:In this work, an evolutionary art project is presented where images are approximated by transparent, overlapping and geometric shapes of different types, e.g., polygons, circles, lines. Genotypes representing features and order of the geometric shapes are evolved with a fitness function that has the corresponding pixels of an input image as a target goal. A genotype-to-phenotype mapping is therefore applied to render images, as the chosen genetic representation is indirect, i.e., genotypes do not include pixels but a combination of shapes with their properties. Different combinations of shapes, quantity of shapes, mutation types and populations are tested. The goal of the work herein is twofold: (1) to approximate images as precisely as possible with evolved indirect encodings, (2) to produce visually appealing results and novel artistic styles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge