Scott J. Rodig
AdvDINO: Domain-Adversarial Self-Supervised Representation Learning for Spatial Proteomics
Aug 07, 2025Abstract:Self-supervised learning (SSL) has emerged as a powerful approach for learning visual representations without manual annotations. However, the robustness of standard SSL methods to domain shift -- systematic differences across data sources -- remains uncertain, posing an especially critical challenge in biomedical imaging where batch effects can obscure true biological signals. We present AdvDINO, a domain-adversarial self-supervised learning framework that integrates a gradient reversal layer into the DINOv2 architecture to promote domain-invariant feature learning. Applied to a real-world cohort of six-channel multiplex immunofluorescence (mIF) whole slide images from non-small cell lung cancer patients, AdvDINO mitigates slide-specific biases to learn more robust and biologically meaningful representations than non-adversarial baselines. Across $>5.46$ million mIF image tiles, the model uncovers phenotype clusters with distinct proteomic profiles and prognostic significance, and improves survival prediction in attention-based multiple instance learning. While demonstrated on mIF data, AdvDINO is broadly applicable to other imaging domains -- including radiology, remote sensing, and autonomous driving -- where domain shift and limited annotated data hinder model generalization and interpretability.
MITI Minimum Information guidelines for highly multiplexed tissue images
Aug 21, 2021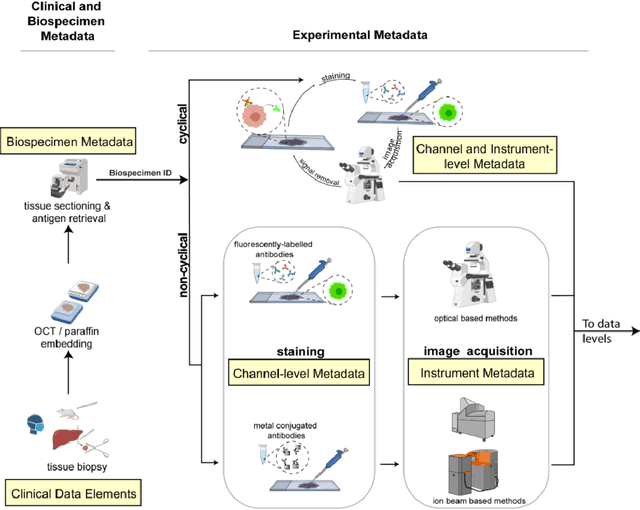
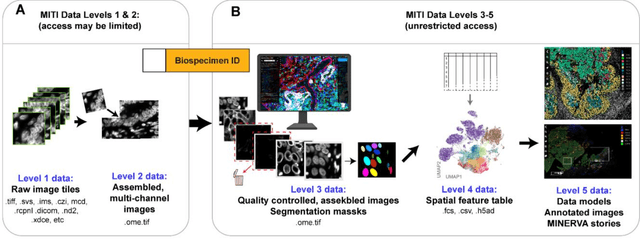
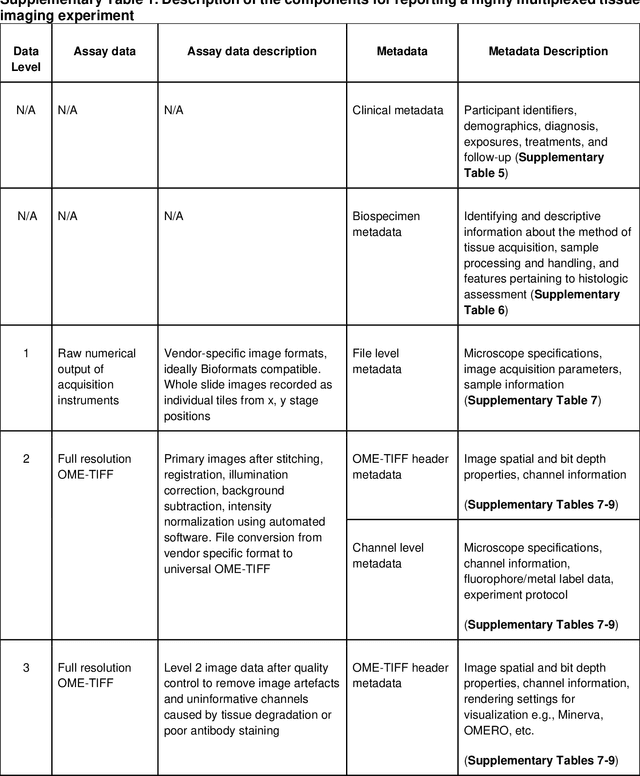
Abstract:The imminent release of atlases combining highly multiplexed tissue imaging with single cell sequencing and other omics data from human tissues and tumors creates an urgent need for data and metadata standards compliant with emerging and traditional approaches to histology. We describe the development of a Minimum Information about highly multiplexed Tissue Imaging (MITI) standard that draws on best practices from genomics and microscopy of cultured cells and model organisms.
Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and Prognosis
Dec 30, 2019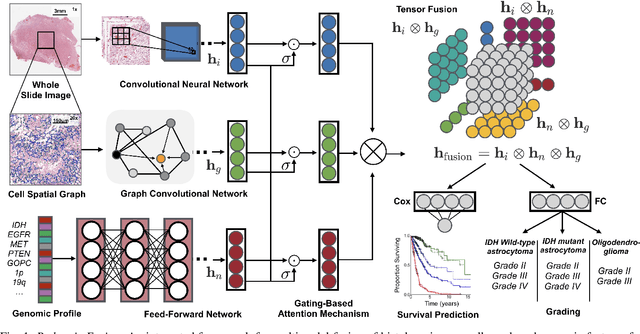
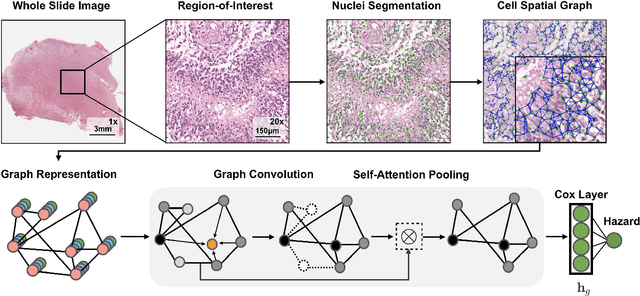
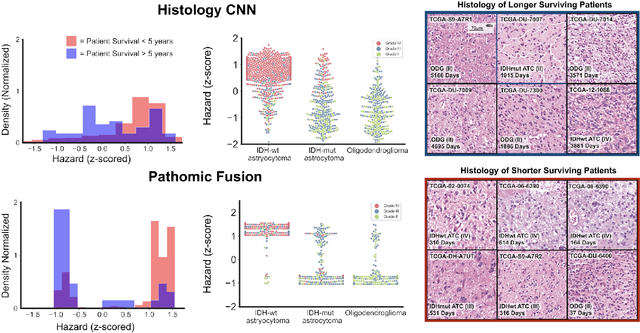
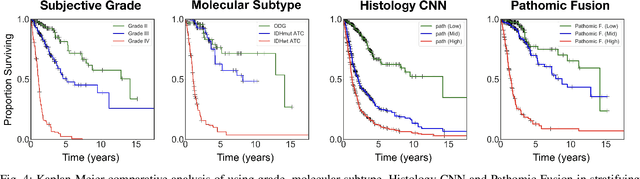
Abstract:Cancer diagnosis, prognosis, and therapeutic response predictions are based on morphological information from histology slides and molecular profiles from genomic data. However, most deep learning-based objective outcome prediction and grading paradigms are based on histology or genomics alone and do not make use of the complementary information in an intuitive manner. In this work, we propose Pathomic Fusion, a strategy for end-to-end multimodal fusion of histology image and genomic (mutations, CNV, mRNAseq) features for survival outcome prediction. Our approach models pairwise feature interactions across modalities by taking the Kronecker product of gated feature representations and controls the expressiveness of each representation via a gating-based attention mechanism. The proposed framework is able to model pairwise interactions across features in different modalities and control their relative importance. We validate our approach using glioma datasets from the Cancer Genome Atlas (TCGA), which contains paired whole-slide image, genotype, and transcriptome data with ground truth survival and histologic grade labels. Based on a rigorous 15-fold cross-validation, our results demonstrate that the proposed multimodal fusion paradigm improves prognostic determinations from grading and molecular subtyping as well as unimodal deep networks trained on histology and genomic data alone. The proposed method establishes insight and theory on how to train deep networks on multimodal biomedical data in an intuitive manner, which will be useful for other problems in medicine that seek to combine heterogeneous data streams for understanding diseases and predicting response and resistance to treatment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge