Richard H. Clayton
Cherise
Seeing Beyond the Image: ECG and Anatomical Knowledge-Guided Myocardial Scar Segmentation from Late Gadolinium-Enhanced Images
Nov 18, 2025



Abstract:Accurate segmentation of myocardial scar from late gadolinium enhanced (LGE) cardiac MRI is essential for evaluating tissue viability, yet remains challenging due to variable contrast and imaging artifacts. Electrocardiogram (ECG) signals provide complementary physiological information, as conduction abnormalities can help localize or suggest scarred myocardial regions. In this work, we propose a novel multimodal framework that integrates ECG-derived electrophysiological information with anatomical priors from the AHA-17 atlas for physiologically consistent LGE-based scar segmentation. As ECGs and LGE-MRIs are not acquired simultaneously, we introduce a Temporal Aware Feature Fusion (TAFF) mechanism that dynamically weights and fuses features based on their acquisition time difference. Our method was evaluated on a clinical dataset and achieved substantial gains over the state-of-the-art image-only baseline (nnU-Net), increasing the average Dice score for scars from 0.6149 to 0.8463 and achieving high performance in both precision (0.9115) and sensitivity (0.9043). These results show that integrating physiological and anatomical knowledge allows the model to "see beyond the image", setting a new direction for robust and physiologically grounded cardiac scar segmentation.
CLAIM: Clinically-Guided LGE Augmentation for Realistic and Diverse Myocardial Scar Synthesis and Segmentation
Jun 18, 2025Abstract:Deep learning-based myocardial scar segmentation from late gadolinium enhancement (LGE) cardiac MRI has shown great potential for accurate and timely diagnosis and treatment planning for structural cardiac diseases. However, the limited availability and variability of LGE images with high-quality scar labels restrict the development of robust segmentation models. To address this, we introduce CLAIM: \textbf{C}linically-Guided \textbf{L}GE \textbf{A}ugmentation for Real\textbf{i}stic and Diverse \textbf{M}yocardial Scar Synthesis and Segmentation framework, a framework for anatomically grounded scar generation and segmentation. At its core is the SMILE module (Scar Mask generation guided by cLinical knowledgE), which conditions a diffusion-based generator on the clinically adopted AHA 17-segment model to synthesize images with anatomically consistent and spatially diverse scar patterns. In addition, CLAIM employs a joint training strategy in which the scar segmentation network is optimized alongside the generator, aiming to enhance both the realism of synthesized scars and the accuracy of the scar segmentation performance. Experimental results show that CLAIM produces anatomically coherent scar patterns and achieves higher Dice similarity with real scar distributions compared to baseline models. Our approach enables controllable and realistic myocardial scar synthesis and has demonstrated utility for downstream medical imaging task.
DenResCov-19: A deep transfer learning network for robust automatic classification of COVID-19, pneumonia, and tuberculosis from X-rays
Apr 08, 2021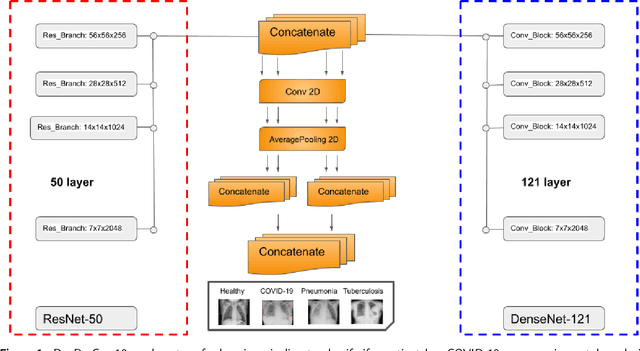
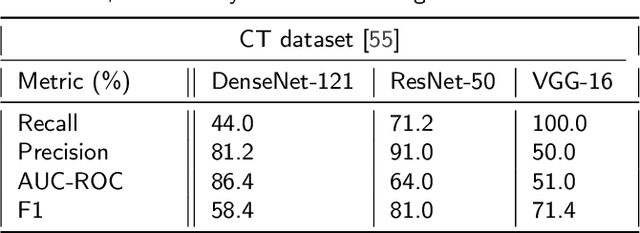
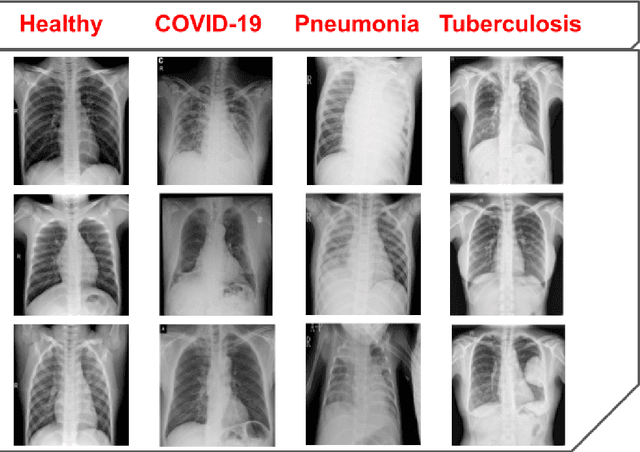
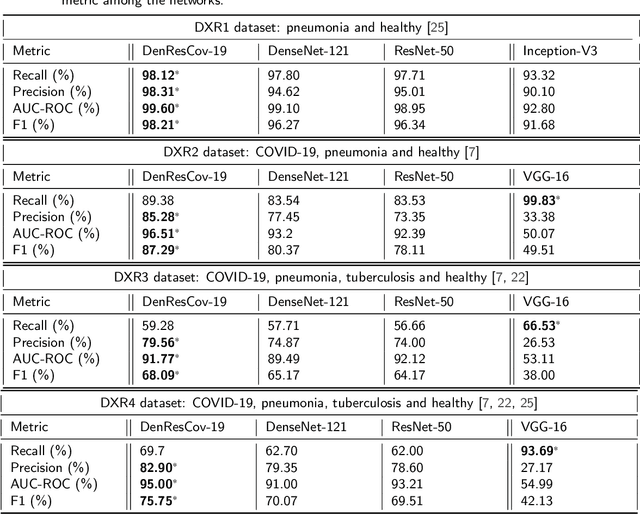
Abstract:The global pandemic of COVID-19 is continuing to have a significant effect on the well-being of global population, increasing the demand for rapid testing, diagnosis, and treatment. Along with COVID-19, other etiologies of pneumonia and tuberculosis constitute additional challenges to the medical system. In this regard, the objective of this work is to develop a new deep transfer learning pipeline to diagnose patients with COVID-19, pneumonia, and tuberculosis, based on chest x-ray images. We observed in some instances DenseNet and Resnet have orthogonal performances. In our proposed model, we have created an extra layer with convolutional neural network blocks to combine these two models to establish superior performance over either model. The same strategy can be useful in other applications where two competing networks with complementary performance are observed. We have tested the performance of our proposed network on two-class (pneumonia vs healthy), three-class (including COVID-19), and four-class (including tuberculosis) classification problems. The proposed network has been able to successfully classify these lung diseases in all four datasets and has provided significant improvement over the benchmark networks of DenseNet, ResNet, and Inception-V3. These novel findings can deliver a state-of-the-art pre-screening fast-track decision network to detect COVID-19 and other lung pathologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge