Paris Perdikaris
Deep learning of free boundary and Stefan problems
Jun 04, 2020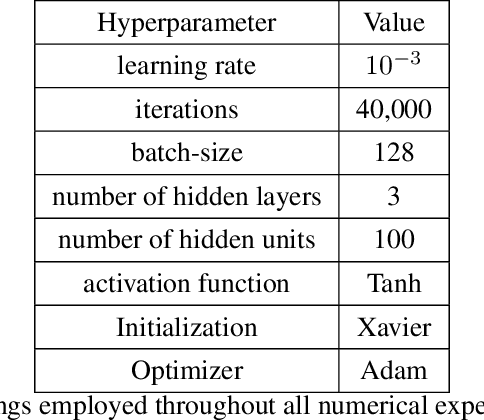
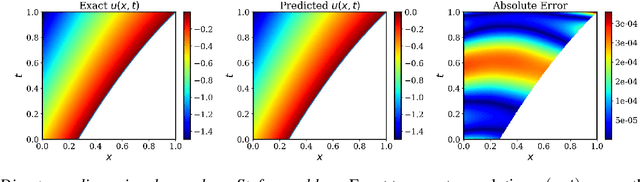
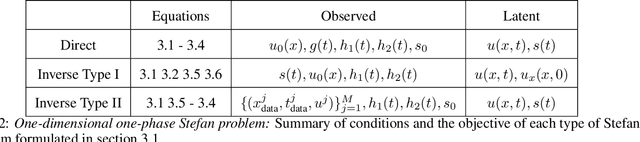
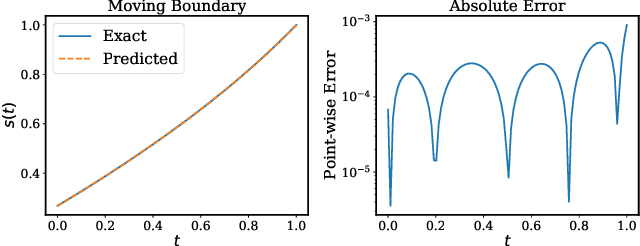
Abstract:Free boundary problems appear naturally in numerous areas of mathematics, science and engineering. These problems present a great computational challenge because they necessitate numerical methods that can yield an accurate approximation of free boundaries and complex dynamic interfaces. In this work, we propose a multi-network model based on physics-informed neural networks to tackle a general class of forward and inverse free boundary problems called Stefan problems. Specifically, we approximate the unknown solution as well as any moving boundaries by two deep neural networks. Besides, we formulate a new type of inverse Stefan problems that aim to reconstruct the solution and free boundaries directly from sparse and noisy measurements. We demonstrate the effectiveness of our approach in a series of benchmarks spanning different types of Stefan problems, and illustrate how the proposed framework can accurately recover solutions of partial differential equations with moving boundaries and dynamic interfaces. All code and data accompanying this manuscript are publicly available at \url{https://github.com/PredictiveIntelligenceLab/DeepStefan}.
Bayesian differential programming for robust systems identification under uncertainty
Apr 18, 2020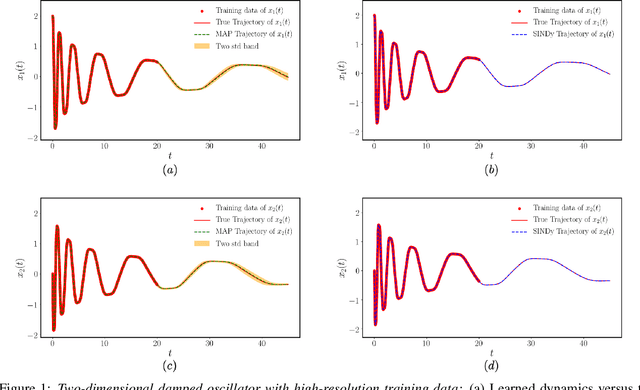
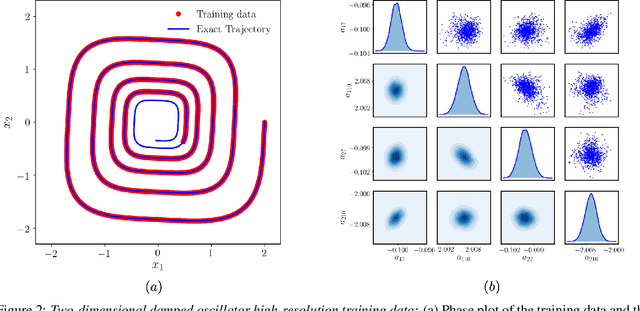
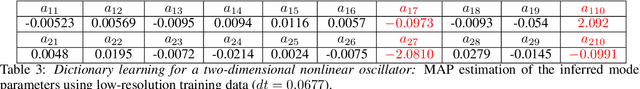
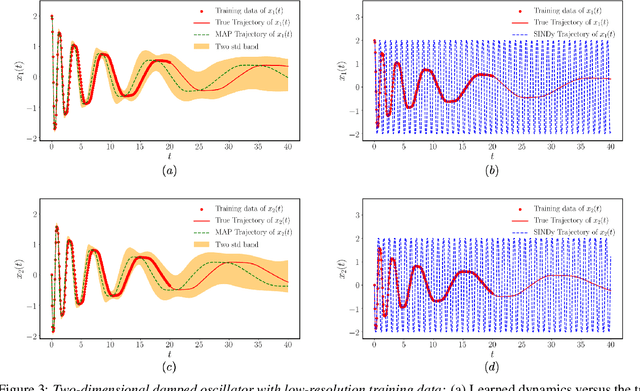
Abstract:This paper presents a machine learning framework for Bayesian systems identification from noisy, sparse and irregular observations of nonlinear dynamical systems. The proposed method takes advantage of recent developments in differentiable programming to propagate gradient information through ordinary differential equation solvers and perform Bayesian inference with respect to unknown model parameters using Hamiltonian Monte Carlo. This allows us to efficiently infer posterior distributions over plausible models with quantified uncertainty, while the use of sparsity-promoting priors enables the discovery of interpretable and parsimonious representations for the underlying latent dynamics. A series of numerical studies is presented to demonstrate the effectiveness of the proposed methods including nonlinear oscillators, predator-prey systems, chaotic dynamics and systems biology. Taken all together, our findings put forth a novel, flexible and robust workflow for data-driven model discovery under uncertainty.
Understanding and mitigating gradient pathologies in physics-informed neural networks
Jan 13, 2020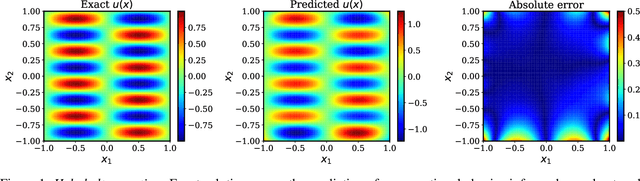
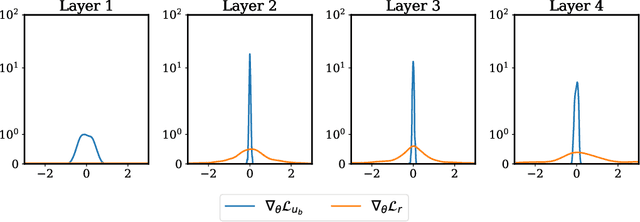
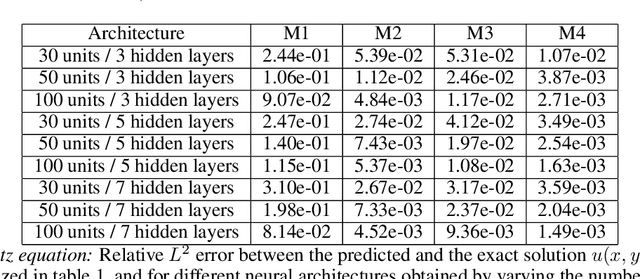
Abstract:The widespread use of neural networks across different scientific domains often involves constraining them to satisfy certain symmetries, conservation laws, or other domain knowledge. Such constraints are often imposed as soft penalties during model training and effectively act as domain-specific regularizers of the empirical risk loss. Physics-informed neural networks is an example of this philosophy in which the outputs of deep neural networks are constrained to approximately satisfy a given set of partial differential equations. In this work we review recent advances in scientific machine learning with a specific focus on the effectiveness of physics-informed neural networks in predicting outcomes of physical systems and discovering hidden physics from noisy data. We will also identify and analyze a fundamental mode of failure of such approaches that is related to numerical stiffness leading to unbalanced back-propagated gradients during model training. To address this limitation we present a learning rate annealing algorithm that utilizes gradient statistics during model training to balance the interplay between different terms in composite loss functions. We also propose a novel neural network architecture that is more resilient to such gradient pathologies. Taken together, our developments provide new insights into the training of constrained neural networks and consistently improve the predictive accuracy of physics-informed neural networks by a factor of 50-100x across a range of problems in computational physics. All code and data accompanying this manuscript are publicly available at \url{https://github.com/PredictiveIntelligenceLab/GradientPathologiesPINNs}.
Machine learning in cardiovascular flows modeling: Predicting pulse wave propagation from non-invasive clinical measurements using physics-informed deep learning
May 13, 2019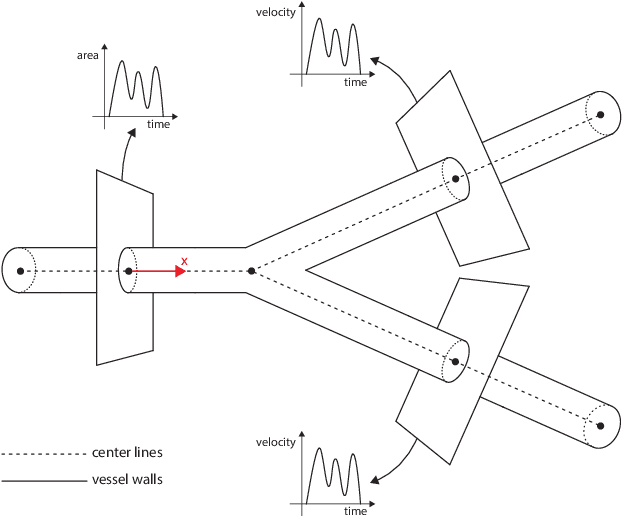
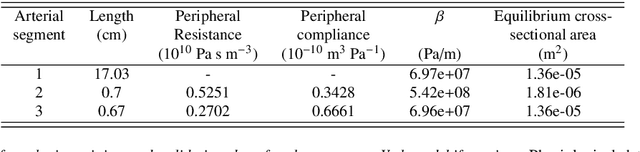
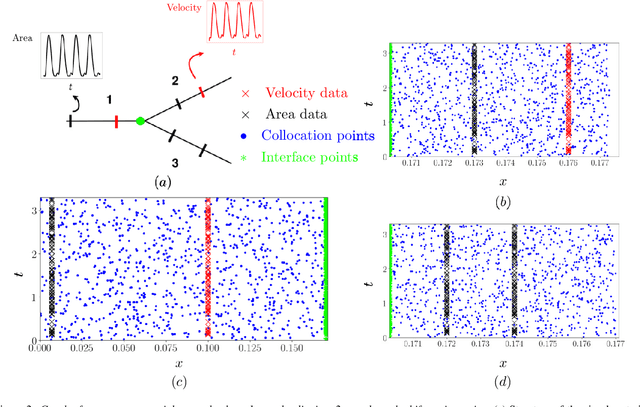
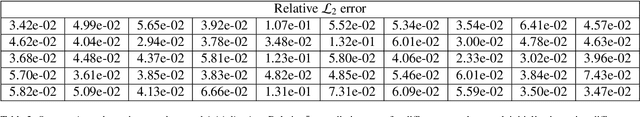
Abstract:Advances in computational science offer a principled pipeline for predictive modeling of cardiovascular flows and aspire to provide a valuable tool for monitoring, diagnostics and surgical planning. Such models can be nowadays deployed on large patient-specific topologies of systemic arterial networks and return detailed predictions on flow patterns, wall shear stresses, and pulse wave propagation. However, their success heavily relies on tedious pre-processing and calibration procedures that typically induce a significant computational cost, thus hampering their clinical applicability. In this work we put forth a machine learning framework that enables the seamless synthesis of non-invasive in-vivo measurement techniques and computational flow dynamics models derived from first physical principles. We illustrate this new paradigm by showing how one-dimensional models of pulsatile flow can be used to constrain the output of deep neural networks such that their predictions satisfy the conservation of mass and momentum principles. Once trained on noisy and scattered clinical data of flow and wall displacement, these networks can return physically consistent predictions for velocity, pressure and wall displacement pulse wave propagation, all without the need to employ conventional simulators. A simple post-processing of these outputs can also provide a cheap and effective way for estimating Windkessel model parameters that are required for the calibration of traditional computational models. The effectiveness of the proposed techniques is demonstrated through a series of prototype benchmarks, as well as a realistic clinical case involving in-vivo measurements near the aorta/carotid bifurcation of a healthy human subject.
Multi-fidelity classification using Gaussian processes: accelerating the prediction of large-scale computational models
May 09, 2019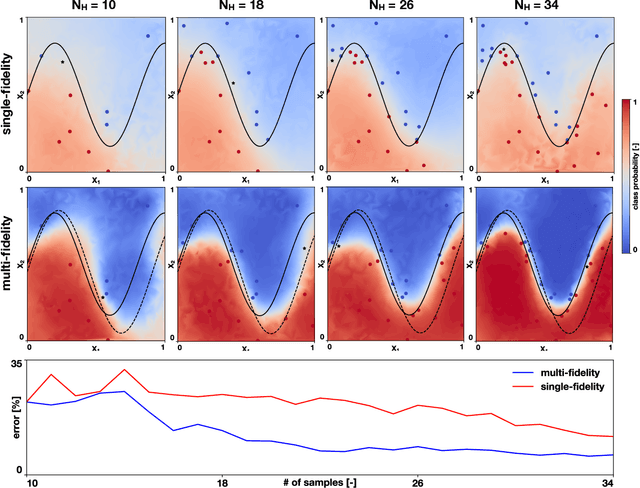
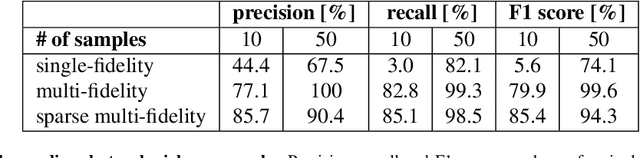
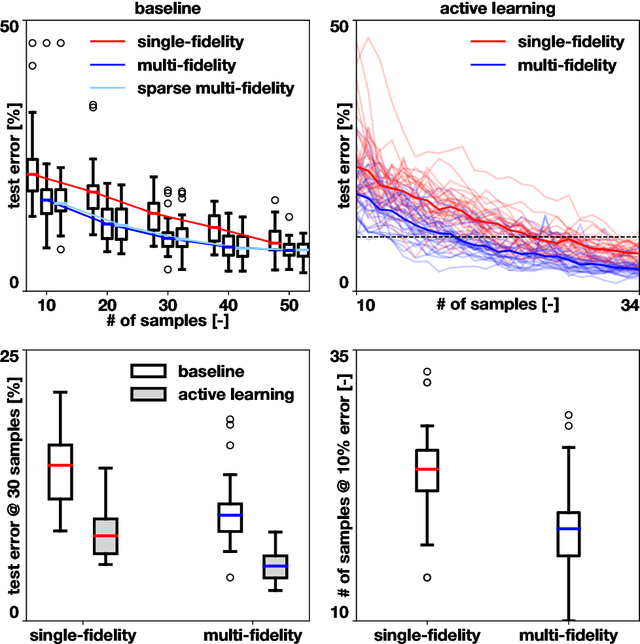
Abstract:Machine learning techniques typically rely on large datasets to create accurate classifiers. However, there are situations when data is scarce and expensive to acquire. This is the case of studies that rely on state-of-the-art computational models which typically take days to run, thus hindering the potential of machine learning tools. In this work, we present a novel classifier that takes advantage of lower fidelity models and inexpensive approximations to predict the binary output of expensive computer simulations. We postulate an autoregressive model between the different levels of fidelity with Gaussian process priors. We adopt a fully Bayesian treatment for the hyper-parameters and use Markov Chain Mont Carlo samplers. We take advantage of the probabilistic nature of the classifier to implement active learning strategies. We also introduce a sparse approximation to enhance the ability of themulti-fidelity classifier to handle large datasets. We test these multi-fidelity classifiers against their single-fidelity counterpart with synthetic data, showing a median computational cost reduction of 23% for a target accuracy of 90%. In an application to cardiac electrophysiology, the multi-fidelity classifier achieves an F1 score, the harmonic mean of precision and recall, of 99.6% compared to 74.1% of a single-fidelity classifier when both are trained with 50 samples. In general, our results show that the multi-fidelity classifiers outperform their single-fidelity counterpart in terms of accuracy in all cases. We envision that this new tool will enable researchers to study classification problems that would otherwise be prohibitively expensive. Source code is available at https://github.com/fsahli/MFclass.
A comparative study of physics-informed neural network models for learning unknown dynamics and constitutive relations
Apr 02, 2019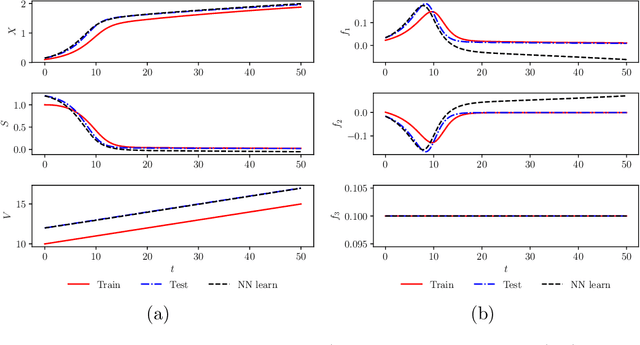
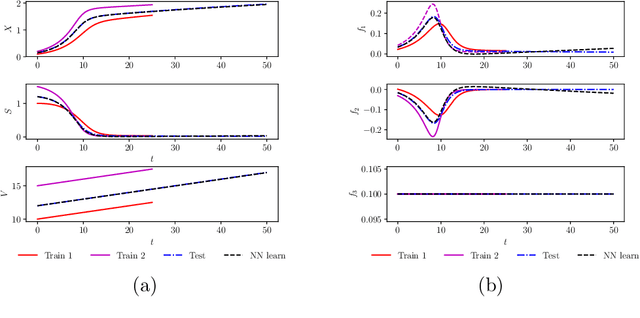
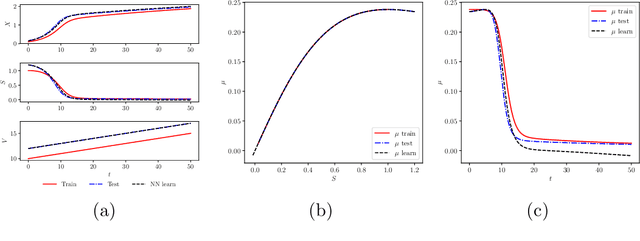
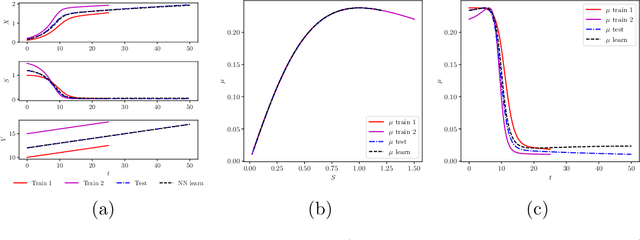
Abstract:We investigate the use of discrete and continuous versions of physics-informed neural network methods for learning unknown dynamics or constitutive relations of a dynamical system. For the case of unknown dynamics, we represent all the dynamics with a deep neural network (DNN). When the dynamics of the system are known up to the specification of constitutive relations (that can depend on the state of the system), we represent these constitutive relations with a DNN. The discrete versions combine classical multistep discretization methods for dynamical systems with neural network based machine learning methods. On the other hand, the continuous versions utilize deep neural networks to minimize the residual function for the continuous governing equations. We use the case of a fedbatch bioreactor system to study the effectiveness of these approaches and discuss conditions for their applicability. Our results indicate that the accuracy of the trained neural network models is much higher for the cases where we only have to learn a constitutive relation instead of the whole dynamics. This finding corroborates the well-known fact from scientific computing that building as much structural information is available into an algorithm can enhance its efficiency and/or accuracy.
Physics-Constrained Deep Learning for High-dimensional Surrogate Modeling and Uncertainty Quantification without Labeled Data
Jan 18, 2019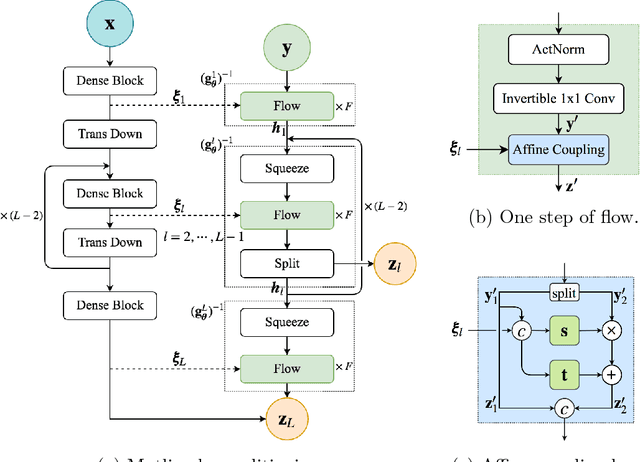
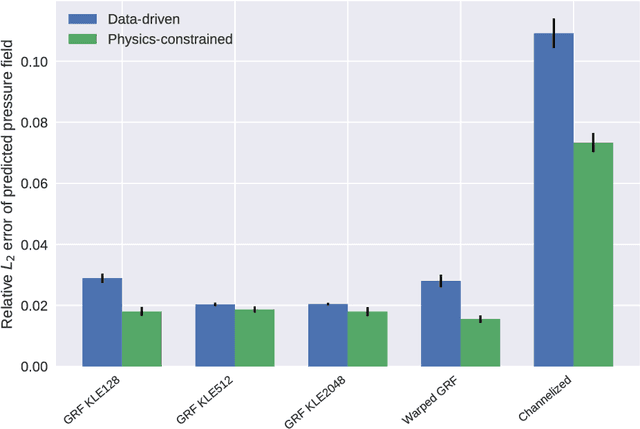
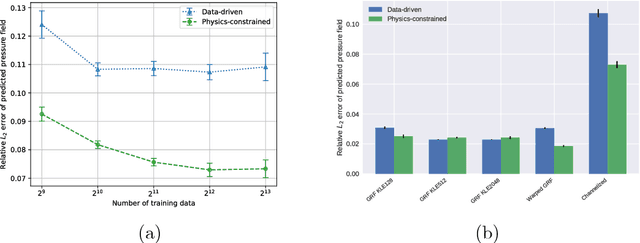
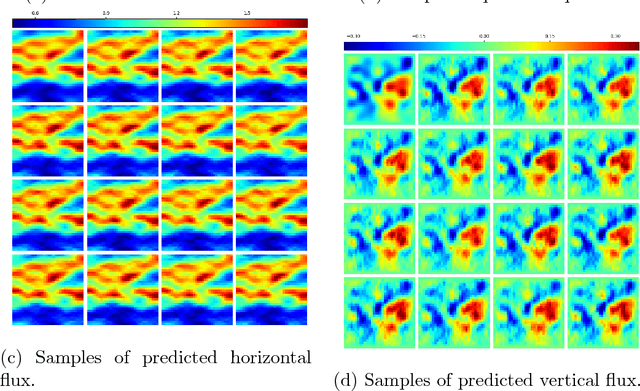
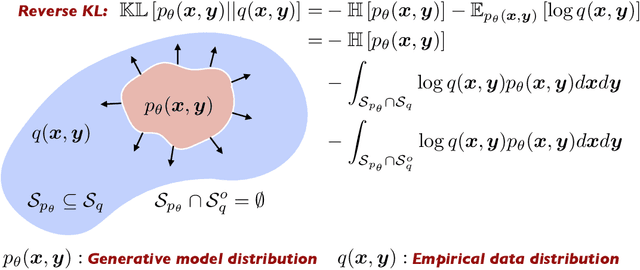
Abstract:Surrogate modeling and uncertainty quantification tasks for PDE systems are most often considered as supervised learning problems where input and output data pairs are used for training. The construction of such emulators is by definition a small data problem which poses challenges to deep learning approaches that have been developed to operate in the big data regime. Even in cases where such models have been shown to have good predictive capability in high dimensions, they fail to address constraints in the data implied by the PDE model. This paper provides a methodology that incorporates the governing equations of the physical model in the loss/likelihood functions. The resulting physics-constrained, deep learning models are trained without any labeled data (e.g. employing only input data) and provide comparable predictive responses with data-driven models while obeying the constraints of the problem at hand. This work employs a convolutional encoder-decoder neural network approach as well as a conditional flow-based generative model for the solution of PDEs, surrogate model construction, and uncertainty quantification tasks. The methodology is posed as a minimization problem of the reverse Kullback-Leibler (KL) divergence between the model predictive density and the reference conditional density, where the later is defined as the Boltzmann-Gibbs distribution at a given inverse temperature with the underlying potential relating to the PDE system of interest. The generalization capability of these models to out-of-distribution input is considered. Quantification and interpretation of the predictive uncertainty is provided for a number of problems.
Conditional deep surrogate models for stochastic, high-dimensional, and multi-fidelity systems
Jan 15, 2019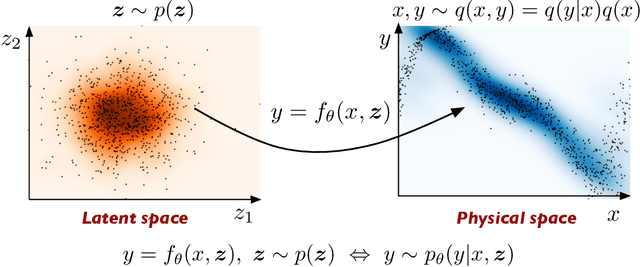
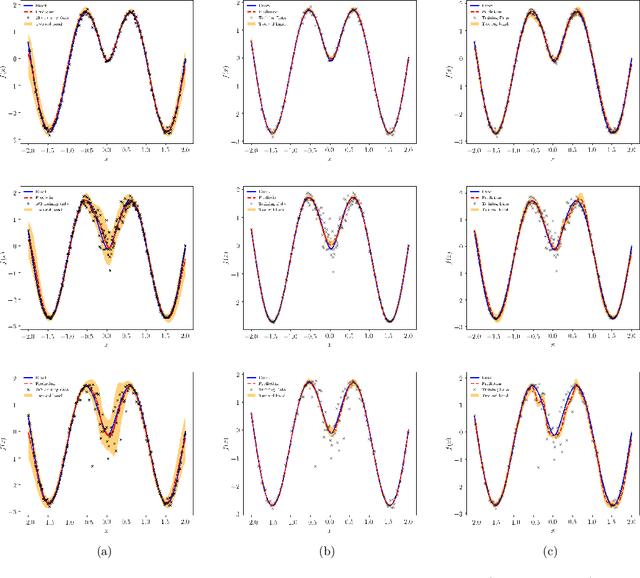
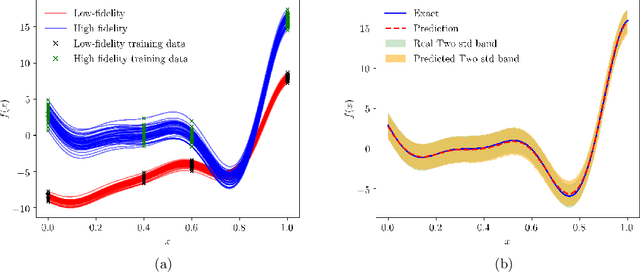
Abstract:We present a probabilistic deep learning methodology that enables the construction of predictive data-driven surrogates for stochastic systems. Leveraging recent advances in variational inference with implicit distributions, we put forth a statistical inference framework that enables the end-to-end training of surrogate models on paired input-output observations that may be stochastic in nature, originate from different information sources of variable fidelity, or be corrupted by complex noise processes. The resulting surrogates can accommodate high-dimensional inputs and outputs and are able to return predictions with quantified uncertainty. The effectiveness our approach is demonstrated through a series of canonical studies, including the regression of noisy data, multi-fidelity modeling of stochastic processes, and uncertainty propagation in high-dimensional dynamical systems.
Physics-informed deep generative models
Dec 09, 2018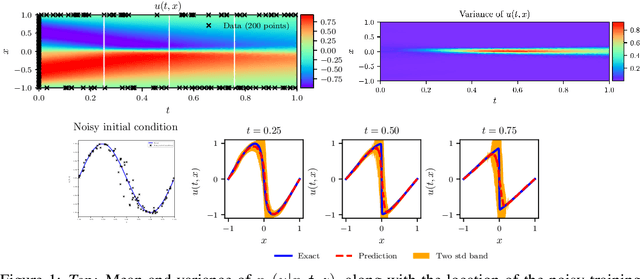


Abstract:We consider the application of deep generative models in propagating uncertainty through complex physical systems. Specifically, we put forth an implicit variational inference formulation that constrains the generative model output to satisfy given physical laws expressed by partial differential equations. Such physics-informed constraints provide a regularization mechanism for effectively training deep probabilistic models for modeling physical systems in which the cost of data acquisition is high and training data-sets are typically small. This provides a scalable framework for characterizing uncertainty in the outputs of physical systems due to randomness in their inputs or noise in their observations. We demonstrate the effectiveness of our approach through a canonical example in transport dynamics.
Adversarial Uncertainty Quantification in Physics-Informed Neural Networks
Nov 09, 2018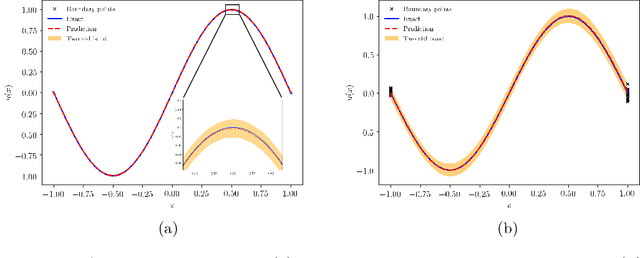
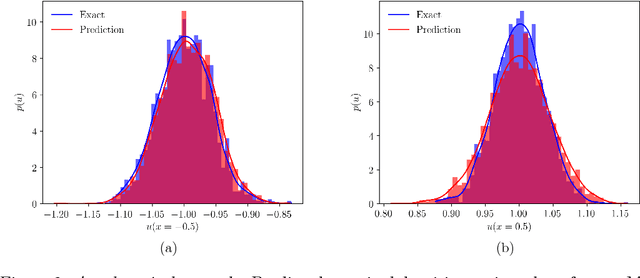
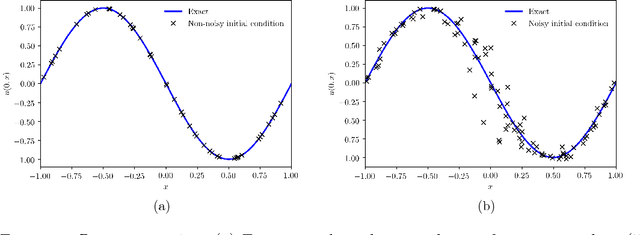
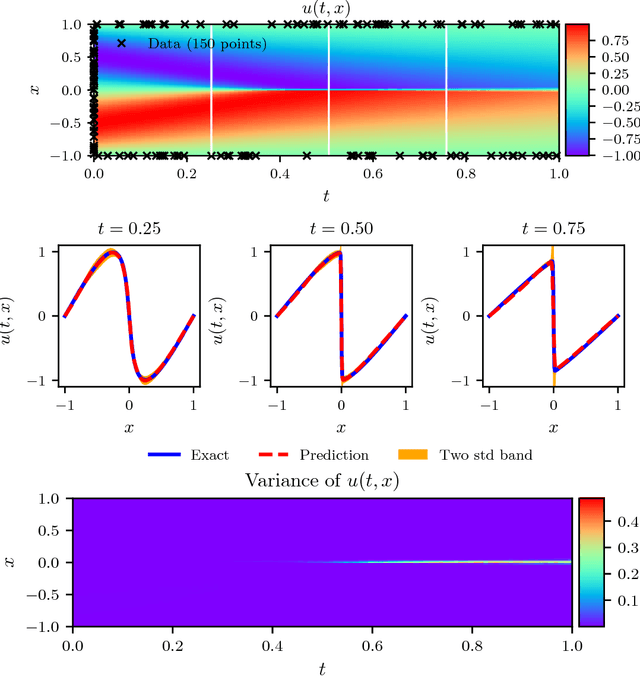
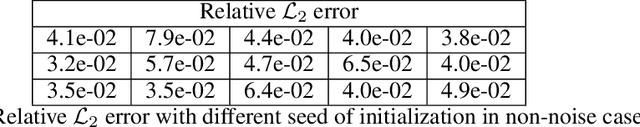
Abstract:We present a deep learning framework for quantifying and propagating uncertainty in systems governed by non-linear differential equations using physics-informed neural networks. Specifically, we employ latent variable models to construct probabilistic representations for the system states, and put forth an adversarial inference procedure for training them on data, while constraining their predictions to satisfy given physical laws expressed by partial differential equations. Such physics-informed constraints provide a regularization mechanism for effectively training deep generative models as surrogates of physical systems in which the cost of data acquisition is high, and training data-sets are typically small. This provides a flexible framework for characterizing uncertainty in the outputs of physical systems due to randomness in their inputs or noise in their observations that entirely bypasses the need for repeatedly sampling expensive experiments or numerical simulators. We demonstrate the effectiveness of our approach through a series of examples involving uncertainty propagation in non-linear conservation laws, and the discovery of constitutive laws for flow through porous media directly from noisy data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge