Ole Solheim
A unified FLAIR hyperintensity segmentation model for various CNS tumor types and acquisition time points
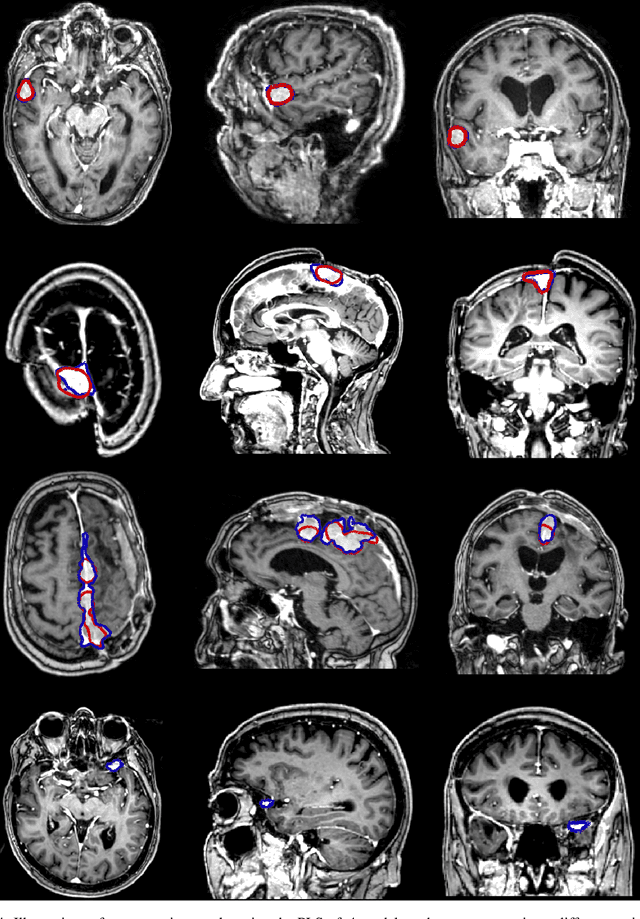
Dec 19, 2025Abstract:T2-weighted fluid-attenuated inversion recovery (FLAIR) magnetic resonance imaging (MRI) scans are important for diagnosis, treatment planning and monitoring of brain tumors. Depending on the brain tumor type, the FLAIR hyperintensity volume is an important measure to asses the tumor volume or surrounding edema, and an automatic segmentation of this would be useful in the clinic. In this study, around 5000 FLAIR images of various tumors types and acquisition time points from different centers were used to train a unified FLAIR hyperintensity segmentation model using an Attention U-Net architecture. The performance was compared against dataset specific models, and was validated on different tumor types, acquisition time points and against BraTS. The unified model achieved an average Dice score of 88.65\% for pre-operative meningiomas, 80.08% for pre-operative metastasis, 90.92% for pre-operative and 84.60% for post-operative gliomas from BraTS, and 84.47% for pre-operative and 61.27\% for post-operative lower grade gliomas. In addition, the results showed that the unified model achieved comparable segmentation performance to the dataset specific models on their respective datasets, and enables generalization across tumor types and acquisition time points, which facilitates the deployment in a clinical setting. The model is integrated into Raidionics, an open-source software for CNS tumor analysis.
Automatic brain tumor segmentation in 2D intra-operative ultrasound images using MRI tumor annotations
Nov 21, 2024



Abstract:Automatic segmentation of brain tumors in intra-operative ultrasound (iUS) images could facilitate localization of tumor tissue during resection surgery. The lack of large annotated datasets limits the current models performances. In this paper, we investigate the use of tumor annotations in pre-operative MRI images, which are more easily accessible than annotations in iUS images, for training of deep learning models for iUS brain tumor segmentation. We used 180 annotated pre-operative MRI images with corresponding unannotated iUS images, and 29 annotated iUS images. Image registration was performed to transfer the MRI annotations to the corresponding iUS images before training models with the nnU-Net framework. To validate the use of MRI labels, the models were compared to a model trained with only US annotated tumors, and a model with both US and MRI annotated tumors. In addition, the results were compared to annotations validated by an expert neurosurgeon on the same test set to measure inter-observer variability. The results showed similar performance for a model trained with only MRI annotated tumors, compared to a model trained with only US annotated tumors. The model trained using both modalities obtained slightly better results with an average Dice score of 0.62, where external expert annotations achieved a score of 0.67. The results also showed that the deep learning models were comparable to expert annotation for larger tumors (> 200 mm2), but perform clearly worse for smaller tumors (< 200 mm2). This shows that MRI tumor annotations can be used as a substitute for US tumor annotations to train a deep learning model for automatic brain tumor segmentation in intra-operative ultrasound images. Small tumors is a limitation for the current models and will be the focus of future work. The main models are available here: https://github.com/mathildefaanes/us_brain_tumor_segmentation.
Postoperative glioblastoma segmentation: Development of a fully automated pipeline using deep convolutional neural networks and comparison with currently available models
Apr 17, 2024



Abstract:Accurately assessing tumor removal is paramount in the management of glioblastoma. We developed a pipeline using MRI scans and neural networks to segment tumor subregions and the surgical cavity in postoperative images. Our model excels in accurately classifying the extent of resection, offering a valuable tool for clinicians in assessing treatment effectiveness.
Segmentation of glioblastomas in early post-operative multi-modal MRI with deep neural networks
Apr 18, 2023Abstract:Extent of resection after surgery is one of the main prognostic factors for patients diagnosed with glioblastoma. To achieve this, accurate segmentation and classification of residual tumor from post-operative MR images is essential. The current standard method for estimating it is subject to high inter- and intra-rater variability, and an automated method for segmentation of residual tumor in early post-operative MRI could lead to a more accurate estimation of extent of resection. In this study, two state-of-the-art neural network architectures for pre-operative segmentation were trained for the task. The models were extensively validated on a multicenter dataset with nearly 1000 patients, from 12 hospitals in Europe and the United States. The best performance achieved was a 61\% Dice score, and the best classification performance was about 80\% balanced accuracy, with a demonstrated ability to generalize across hospitals. In addition, the segmentation performance of the best models was on par with human expert raters. The predicted segmentations can be used to accurately classify the patients into those with residual tumor, and those with gross total resection.
RESECT-SEG: Open access annotations of intra-operative brain tumor ultrasound images
Jul 13, 2022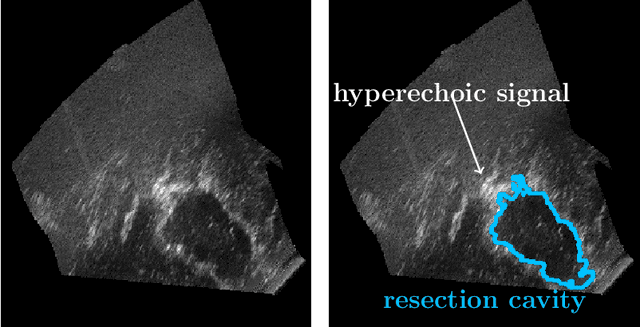
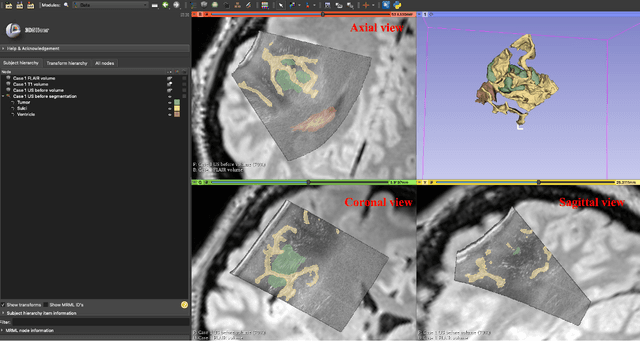
Abstract:Purpose: Registration and segmentation of magnetic resonance (MR) and ultrasound (US) images play an essential role in surgical planning and resection of brain tumors. However, validating these techniques is challenging due to the scarcity of publicly accessible sources with high-quality ground truth information. To this end, we propose a unique annotation dataset of tumor tissues and resection cavities from the previously published RESECT dataset (Xiao et al. 2017) to encourage a more rigorous assessments of image processing techniques. Acquisition and validation methods: The RESECT database consists of MR and intraoperative US (iUS) images of 23 patients who underwent resection surgeries. The proposed dataset contains tumor tissues and resection cavity annotations of the iUS images. The quality of annotations were validated by two highly experienced neurosurgeons through several assessment criteria. Data format and availability: Annotations of tumor tissues and resection cavities are provided in 3D NIFTI formats. Both sets of annotations are accessible online in the \url{https://osf.io/6y4db}. Discussion and potential applications: The proposed database includes tumor tissue and resection cavity annotations from real-world clinical ultrasound brain images to evaluate segmentation and registration methods. These labels could also be used to train deep learning approaches. Eventually, this dataset should further improve the quality of image guidance in neurosurgery.
Meningioma segmentation in T1-weighted MRI leveraging global context and attention mechanisms
Jan 19, 2021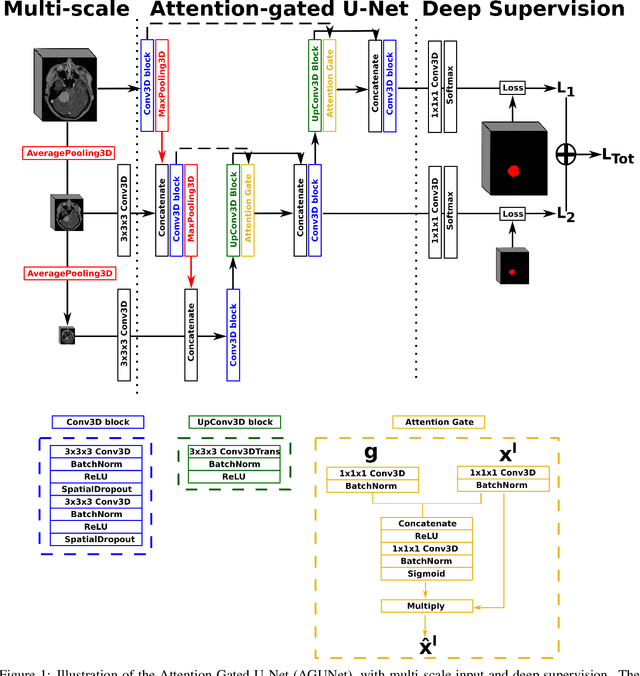
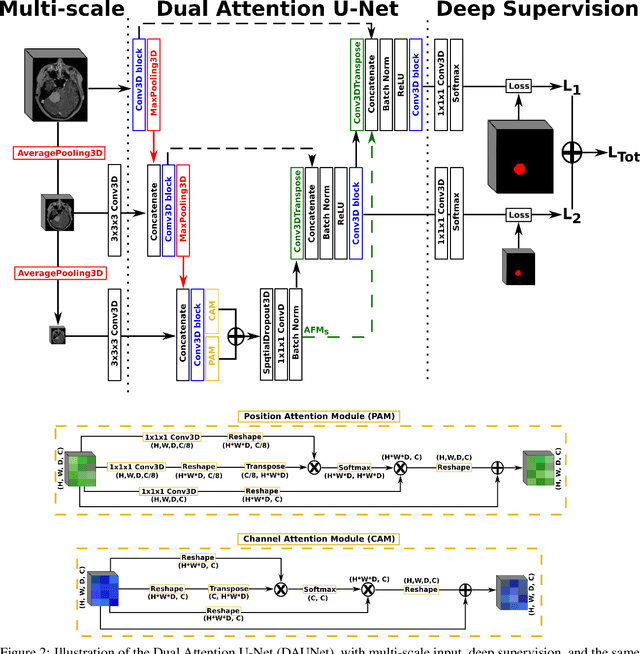
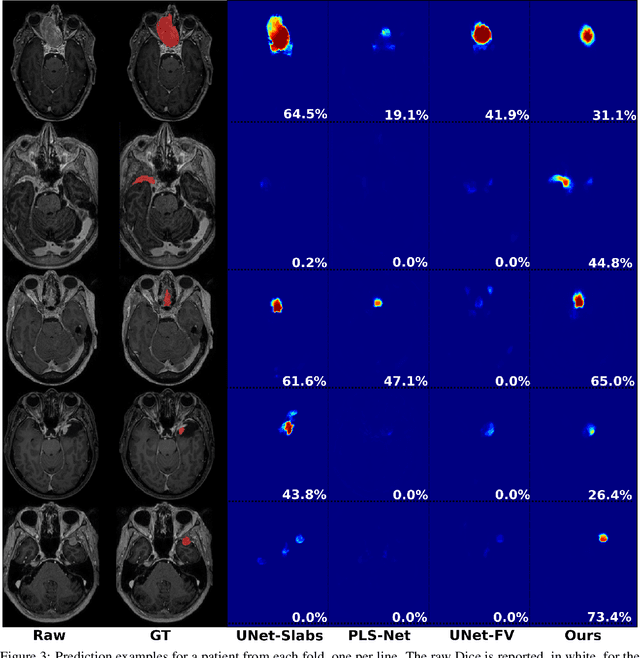
Abstract:Meningiomas are the most common type of primary brain tumor, accounting for approximately 30% of all brain tumors. A substantial number of these tumors are never surgically removed but rather monitored over time. Automatic and precise meningioma segmentation is therefore beneficial to enable reliable growth estimation and patient-specific treatment planning. In this study, we propose the inclusion of attention mechanisms over a U-Net architecture: (i) Attention-gated U-Net (AGUNet) and (ii) Dual Attention U-Net (DAUNet), using a 3D MRI volume as input. Attention has the potential to leverage the global context and identify features' relationships across the entire volume. To limit spatial resolution degradation and loss of detail inherent to encoder-decoder architectures, we studied the impact of multi-scale input and deep supervision components. The proposed architectures are trainable end-to-end and each concept can be seamlessly disabled for ablation studies. The validation studies were performed using a 5-fold cross validation over 600 T1-weighted MRI volumes from St. Olavs University Hospital, Trondheim, Norway. For the best performing architecture, an average Dice score of 81.6% was reached for an F1-score of 95.6%. With an almost perfect precision of 98%, meningiomas smaller than 3ml were occasionally missed hence reaching an overall recall of 93%. Leveraging global context from a 3D MRI volume provided the best performances, even if the native volume resolution could not be processed directly. Overall, near-perfect detection was achieved for meningiomas larger than 3ml which is relevant for clinical use. In the future, the use of multi-scale designs and refinement networks should be further investigated to improve the performance. A larger number of cases with meningiomas below 3ml might also be needed to improve the performance for the smallest tumors.
Fast meningioma segmentation in T1-weighted MRI volumes using a lightweight 3D deep learning architecture
Oct 14, 2020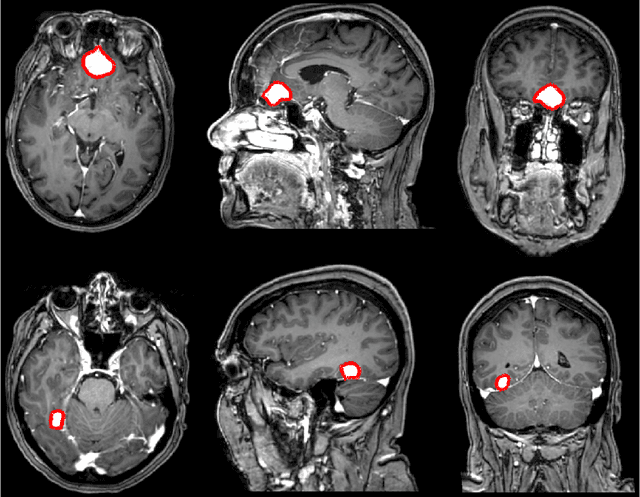
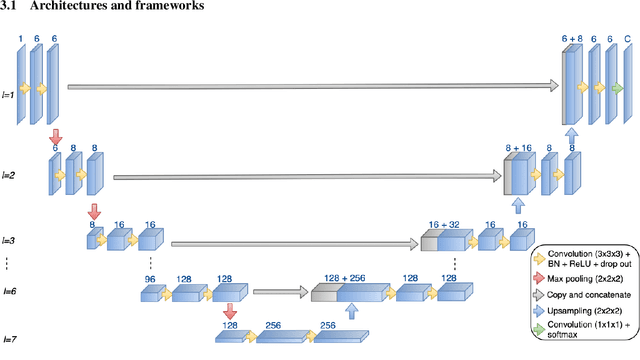
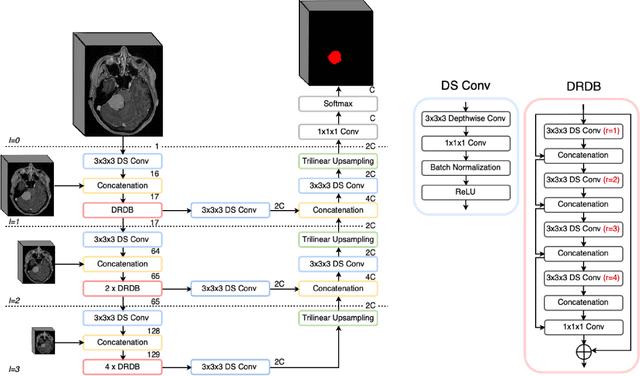
Abstract:Automatic and consistent meningioma segmentation in T1-weighted MRI volumes and corresponding volumetric assessment is of use for diagnosis, treatment planning, and tumor growth evaluation. In this paper, we optimized the segmentation and processing speed performances using a large number of both surgically treated meningiomas and untreated meningiomas followed at the outpatient clinic. We studied two different 3D neural network architectures: (i) a simple encoder-decoder similar to a 3D U-Net, and (ii) a lightweight multi-scale architecture (PLS-Net). In addition, we studied the impact of different training schemes. For the validation studies, we used 698 T1-weighted MR volumes from St. Olav University Hospital, Trondheim, Norway. The models were evaluated in terms of detection accuracy, segmentation accuracy and training/inference speed. While both architectures reached a similar Dice score of 70% on average, the PLS-Net was more accurate with an F1-score of up to 88%. The highest accuracy was achieved for the largest meningiomas. Speed-wise, the PLS-Net architecture tended to converge in about 50 hours while 130 hours were necessary for U-Net. Inference with PLS-Net takes less than a second on GPU and about 15 seconds on CPU. Overall, with the use of mixed precision training, it was possible to train competitive segmentation models in a relatively short amount of time using the lightweight PLS-Net architecture. In the future, the focus should be brought toward the segmentation of small meningiomas (less than 2ml) to improve clinical relevance for automatic and early diagnosis as well as speed of growth estimates.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge