Nathan Gaw
Uni-3DAD: GAN-Inversion Aided Universal 3D Anomaly Detection on Model-free Products
Aug 29, 2024



Abstract:Anomaly detection is a long-standing challenge in manufacturing systems. Traditionally, anomaly detection has relied on human inspectors. However, 3D point clouds have gained attention due to their robustness to environmental factors and their ability to represent geometric data. Existing 3D anomaly detection methods generally fall into two categories. One compares scanned 3D point clouds with design files, assuming these files are always available. However, such assumptions are often violated in many real-world applications where model-free products exist, such as fresh produce (i.e., ``Cookie", ``Potato", etc.), dentures, bone, etc. The other category compares patches of scanned 3D point clouds with a library of normal patches named memory bank. However, those methods usually fail to detect incomplete shapes, which is a fairly common defect type (i.e., missing pieces of different products). The main challenge is that missing areas in 3D point clouds represent the absence of scanned points. This makes it infeasible to compare the missing region with existing point cloud patches in the memory bank. To address these two challenges, we proposed a unified, unsupervised 3D anomaly detection framework capable of identifying all types of defects on model-free products. Our method integrates two detection modules: a feature-based detection module and a reconstruction-based detection module. Feature-based detection covers geometric defects, such as dents, holes, and cracks, while the reconstruction-based method detects missing regions. Additionally, we employ a One-class Support Vector Machine (OCSVM) to fuse the detection results from both modules. The results demonstrate that (1) our proposed method outperforms the state-of-the-art methods in identifying incomplete shapes and (2) it still maintains comparable performance with the SOTA methods in detecting all other types of anomalies.
H2G2-Net: A Hierarchical Heterogeneous Graph Generative Network Framework for Discovery of Multi-Modal Physiological Responses
Jan 05, 2024Abstract:Discovering human cognitive and emotional states using multi-modal physiological signals draws attention across various research applications. Physiological responses of the human body are influenced by human cognition and commonly used to analyze cognitive states. From a network science perspective, the interactions of these heterogeneous physiological modalities in a graph structure may provide insightful information to support prediction of cognitive states. However, there is no clue to derive exact connectivity between heterogeneous modalities and there exists a hierarchical structure of sub-modalities. Existing graph neural networks are designed to learn on non-hierarchical homogeneous graphs with pre-defined graph structures; they failed to learn from hierarchical, multi-modal physiological data without a pre-defined graph structure. To this end, we propose a hierarchical heterogeneous graph generative network (H2G2-Net) that automatically learns a graph structure without domain knowledge, as well as a powerful representation on the hierarchical heterogeneous graph in an end-to-end fashion. We validate the proposed method on the CogPilot dataset that consists of multi-modal physiological signals. Extensive experiments demonstrate that our proposed method outperforms the state-of-the-art GNNs by 5%-20% in prediction accuracy.
Assessing the (Un)Trustworthiness of Saliency Maps for Localizing Abnormalities in Medical Imaging
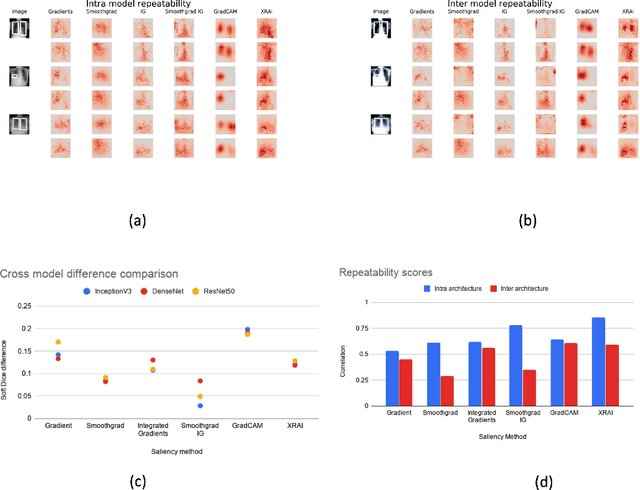
Aug 06, 2020Abstract:Saliency maps have become a widely used method to make deep learning models more interpretable by providing post-hoc explanations of classifiers through identification of the most pertinent areas of the input medical image. They are increasingly being used in medical imaging to provide clinically plausible explanations for the decisions the neural network makes. However, the utility and robustness of these visualization maps has not yet been rigorously examined in the context of medical imaging. We posit that trustworthiness in this context requires 1) localization utility, 2) sensitivity to model weight randomization, 3) repeatability, and 4) reproducibility. Using the localization information available in two large public radiology datasets, we quantify the performance of eight commonly used saliency map approaches for the above criteria using area under the precision-recall curves (AUPRC) and structural similarity index (SSIM), comparing their performance to various baseline measures. Using our framework to quantify the trustworthiness of saliency maps, we show that all eight saliency map techniques fail at least one of the criteria and are, in most cases, less trustworthy when compared to the baselines. We suggest that their usage in the high-risk domain of medical imaging warrants additional scrutiny and recommend that detection or segmentation models be used if localization is the desired output of the network. Additionally, to promote reproducibility of our findings, we provide the code we used for all tests performed in this work at this link: https://github.com/QTIM-Lab/Assessing-Saliency-Maps.
Assessing the validity of saliency maps for abnormality localization in medical imaging
May 29, 2020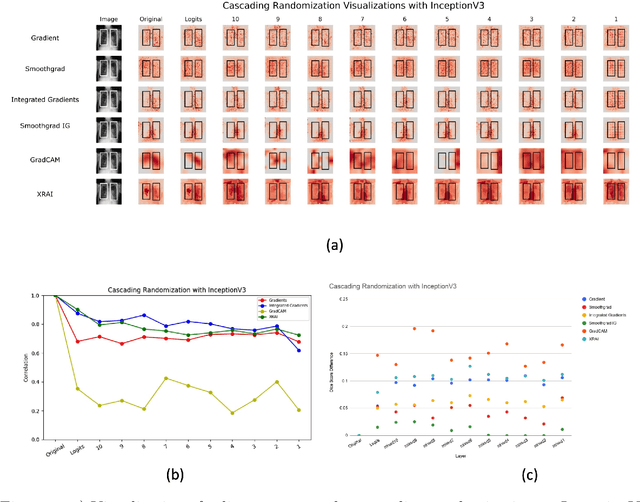
Abstract:Saliency maps have become a widely used method to assess which areas of the input image are most pertinent to the prediction of a trained neural network. However, in the context of medical imaging, there is no study to our knowledge that has examined the efficacy of these techniques and quantified them using overlap with ground truth bounding boxes. In this work, we explored the credibility of the various existing saliency map methods on the RSNA Pneumonia dataset. We found that GradCAM was the most sensitive to model parameter and label randomization, and was highly agnostic to model architecture.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge