Mustaffa Hussain
Switched auxiliary loss for robust training of transformer models for histopathological image segmentation
Aug 21, 2023



Abstract:Functional tissue Units (FTUs) are cell population neighborhoods local to a particular organ performing its main function. The FTUs provide crucial information to the pathologist in understanding the disease affecting a particular organ by providing information at the cellular level. In our research, we have developed a model to segment multi-organ FTUs across 5 organs namely: the kidney, large intestine, lung, prostate and spleen by utilizing the HuBMAP + HPA - Hacking the Human Body competition dataset. We propose adding shifted auxiliary loss for training models like the transformers to overcome the diminishing gradient problem which poses a challenge towards optimal training of deep models. Overall, our model achieved a dice score of 0.793 on the public dataset and 0.778 on the private dataset and shows a 1% improvement with the use of the proposed method. The findings also bolster the use of transformers models for dense prediction tasks in the field of medical image analysis. The study assists in understanding the relationships between cell and tissue organization thereby providing a useful medium to look at the impact of cellular functions on human health.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Robust Multi-Domain Mitosis Detection
Sep 13, 2021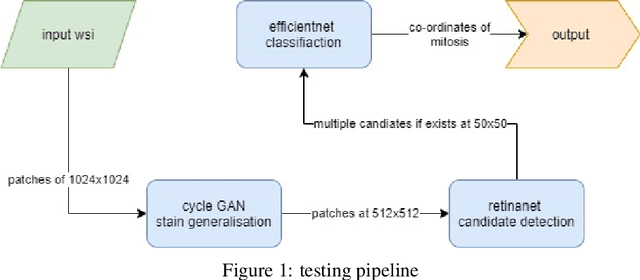
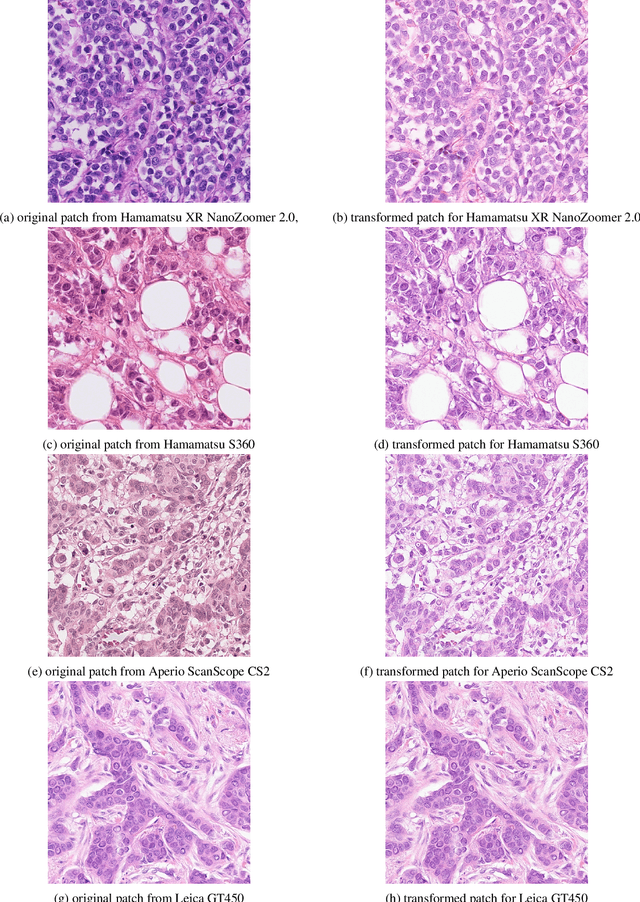
Abstract:Domain variability is a common bottle neck in developing generalisable algorithms for various medical applications. Motivated by the observation that the domain variability of the medical images is to some extent compact, we propose to learn a target representative feature space through unpaired image to image translation (CycleGAN). We comprehensively evaluate the performanceand usefulness by utilising the transformation to mitosis detection with candidate proposal and classification. This work presents a simple yet effective multi-step mitotic figure detection algorithm developed as a baseline for the MIDOG challenge. On the preliminary test set, the algorithm scoresan F1 score of 0.52.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge