Miriam Hägele
DiffInfinite: Large Mask-Image Synthesis via Parallel Random Patch Diffusion in Histopathology
Jun 23, 2023



Abstract:We present DiffInfinite, a hierarchical diffusion model that generates arbitrarily large histological images while preserving long-range correlation structural information. Our approach first generates synthetic segmentation masks, subsequently used as conditions for the high-fidelity generative diffusion process. The proposed sampling method can be scaled up to any desired image size while only requiring small patches for fast training. Moreover, it can be parallelized more efficiently than previous large-content generation methods while avoiding tiling artefacts. The training leverages classifier-free guidance to augment a small, sparsely annotated dataset with unlabelled data. Our method alleviates unique challenges in histopathological imaging practice: large-scale information, costly manual annotation, and protective data handling. The biological plausibility of DiffInfinite data is validated in a survey by ten experienced pathologists as well as a downstream segmentation task. Furthermore, the model scores strongly on anti-copying metrics which is beneficial for the protection of patient data.
Leveraging weak complementary labels to improve semantic segmentation of hepatocellular carcinoma and cholangiocarcinoma in H&E-stained slides
Feb 03, 2023



Abstract:In this paper, we present a deep learning segmentation approach to classify and quantify the two most prevalent primary liver cancers - hepatocellular carcinoma and intrahepatic cholangiocarcinoma - from hematoxylin and eosin (H&E) stained whole slide images. While semantic segmentation of medical images typically requires costly pixel-level annotations by domain experts, there often exists additional information which is routinely obtained in clinical diagnostics but rarely utilized for model training. We propose to leverage such weak information from patient diagnoses by deriving complementary labels that indicate to which class a sample cannot belong to. To integrate these labels, we formulate a complementary loss for segmentation. Motivated by the medical application, we demonstrate for general segmentation tasks that including additional patches with solely weak complementary labels during model training can significantly improve the predictive performance and robustness of a model. On the task of diagnostic differentiation between hepatocellular carcinoma and intrahepatic cholangiocarcinoma, we achieve a balanced accuracy of 0.91 (CI 95%: 0.86 - 0.95) at case level for 165 hold-out patients. Furthermore, we also show that leveraging complementary labels improves the robustness of segmentation and increases performance at case level.
Resolving challenges in deep learning-based analyses of histopathological images using explanation methods
Aug 15, 2019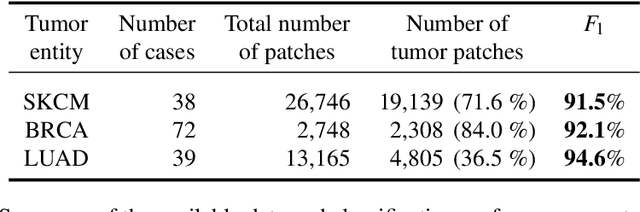
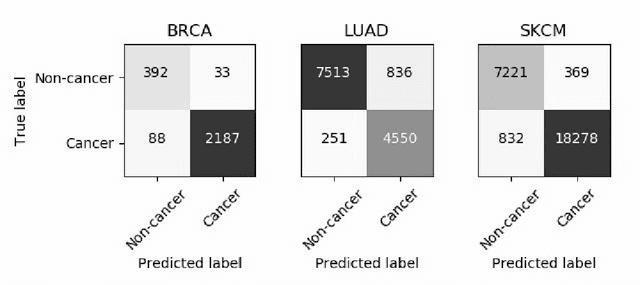
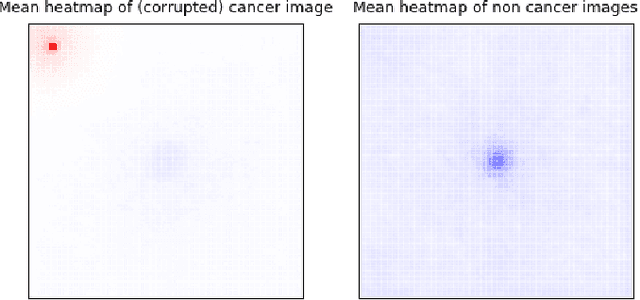
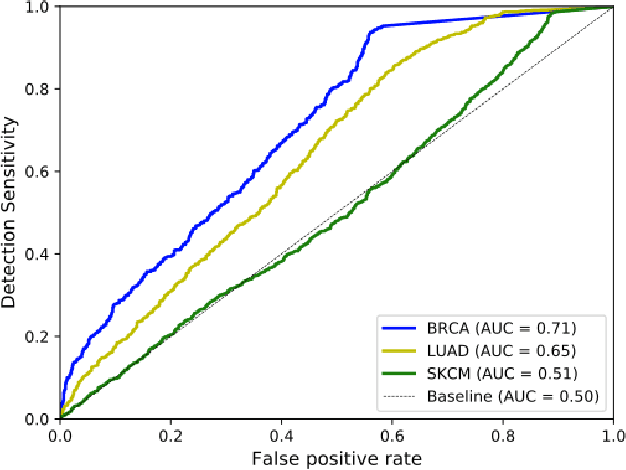
Abstract:Deep learning has recently gained popularity in digital pathology due to its high prediction quality. However, the medical domain requires explanation and insight for a better understanding beyond standard quantitative performance evaluation. Recently, explanation methods have emerged, which are so far still rarely used in medicine. This work shows their application to generate heatmaps that allow to resolve common challenges encountered in deep learning-based digital histopathology analyses. These challenges comprise biases typically inherent to histopathology data. We study binary classification tasks of tumor tissue discrimination in publicly available haematoxylin and eosin slides of various tumor entities and investigate three types of biases: (1) biases which affect the entire dataset, (2) biases which are by chance correlated with class labels and (3) sampling biases. While standard analyses focus on patch-level evaluation, we advocate pixel-wise heatmaps, which offer a more precise and versatile diagnostic instrument and furthermore help to reveal biases in the data. This insight is shown to not only detect but also to be helpful to remove the effects of common hidden biases, which improves generalization within and across datasets. For example, we could see a trend of improved area under the receiver operating characteristic curve by 5% when reducing a labeling bias. Explanation techniques are thus demonstrated to be a helpful and highly relevant tool for the development and the deployment phases within the life cycle of real-world applications in digital pathology.
iNNvestigate neural networks!
Aug 13, 2018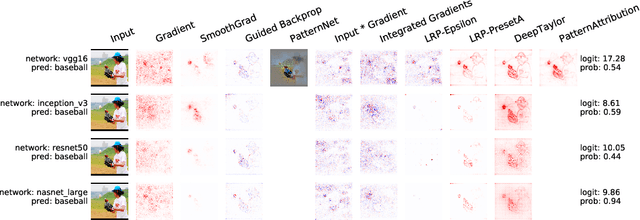
Abstract:In recent years, deep neural networks have revolutionized many application domains of machine learning and are key components of many critical decision or predictive processes. Therefore, it is crucial that domain specialists can understand and analyze actions and pre- dictions, even of the most complex neural network architectures. Despite these arguments neural networks are often treated as black boxes. In the attempt to alleviate this short- coming many analysis methods were proposed, yet the lack of reference implementations often makes a systematic comparison between the methods a major effort. The presented library iNNvestigate addresses this by providing a common interface and out-of-the- box implementation for many analysis methods, including the reference implementation for PatternNet and PatternAttribution as well as for LRP-methods. To demonstrate the versatility of iNNvestigate, we provide an analysis of image classifications for variety of state-of-the-art neural network architectures.
Towards computational fluorescence microscopy: Machine learning-based integrated prediction of morphological and molecular tumor profiles
May 28, 2018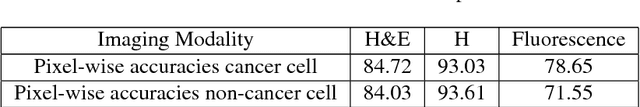
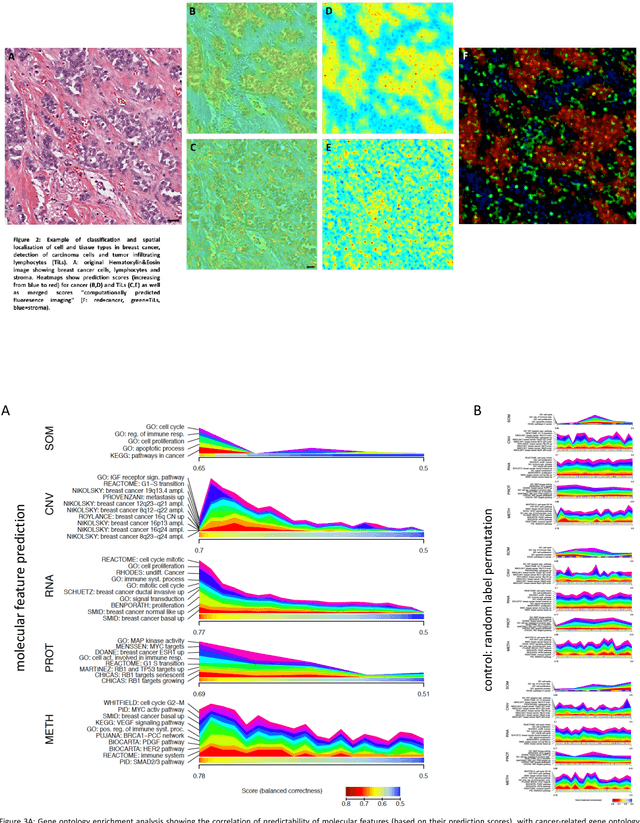

Abstract:Recent advances in cancer research largely rely on new developments in microscopic or molecular profiling techniques offering high level of detail with respect to either spatial or molecular features, but usually not both. Here, we present a novel machine learning-based computational approach that allows for the identification of morphological tissue features and the prediction of molecular properties from breast cancer imaging data. This integration of microanatomic information of tumors with complex molecular profiling data, including protein or gene expression, copy number variation, gene methylation and somatic mutations, provides a novel means to computationally score molecular markers with respect to their relevance to cancer and their spatial associations within the tumor microenvironment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge