Mingxuan Gu
Focus on Content not Noise: Improving Image Generation for Nuclei Segmentation by Suppressing Steganography in CycleGAN
Aug 03, 2023Abstract:Annotating nuclei in microscopy images for the training of neural networks is a laborious task that requires expert knowledge and suffers from inter- and intra-rater variability, especially in fluorescence microscopy. Generative networks such as CycleGAN can inverse the process and generate synthetic microscopy images for a given mask, thereby building a synthetic dataset. However, past works report content inconsistencies between the mask and generated image, partially due to CycleGAN minimizing its loss by hiding shortcut information for the image reconstruction in high frequencies rather than encoding the desired image content and learning the target task. In this work, we propose to remove the hidden shortcut information, called steganography, from generated images by employing a low pass filtering based on the DCT. We show that this increases coherence between generated images and cycled masks and evaluate synthetic datasets on a downstream nuclei segmentation task. Here we achieve an improvement of 5.4 percentage points in the F1-score compared to a vanilla CycleGAN. Integrating advanced regularization techniques into the CycleGAN architecture may help mitigate steganography-related issues and produce more accurate synthetic datasets for nuclei segmentation.
Motion Compensation via Epipolar Consistency for In-Vivo X-Ray Microscopy
Mar 01, 2023

Abstract:Intravital X-ray microscopy (XRM) in preclinical mouse models is of vital importance for the identification of microscopic structural pathological changes in the bone which are characteristic of osteoporosis. The complexity of this method stems from the requirement for high-quality 3D reconstructions of the murine bones. However, respiratory motion and muscle relaxation lead to inconsistencies in the projection data which result in artifacts in uncompensated reconstructions. Motion compensation using epipolar consistency conditions (ECC) has previously shown good performance in clinical CT settings. Here, we explore whether such algorithms are suitable for correcting motion-corrupted XRM data. Different rigid motion patterns are simulated and the quality of the motion-compensated reconstructions is assessed. The method is able to restore microscopic features for out-of-plane motion, but artifacts remain for more realistic motion patterns including all six degrees of freedom of rigid motion. Therefore, ECC is valuable for the initial alignment of the projection data followed by further fine-tuning of motion parameters using a reconstruction-based method
Noise2Contrast: Multi-Contrast Fusion Enables Self-Supervised Tomographic Image Denoising
Dec 09, 2022



Abstract:Self-supervised image denoising techniques emerged as convenient methods that allow training denoising models without requiring ground-truth noise-free data. Existing methods usually optimize loss metrics that are calculated from multiple noisy realizations of similar images, e.g., from neighboring tomographic slices. However, those approaches fail to utilize the multiple contrasts that are routinely acquired in medical imaging modalities like MRI or dual-energy CT. In this work, we propose the new self-supervised training scheme Noise2Contrast that combines information from multiple measured image contrasts to train a denoising model. We stack denoising with domain-transfer operators to utilize the independent noise realizations of different image contrasts to derive a self-supervised loss. The trained denoising operator achieves convincing quantitative and qualitative results, outperforming state-of-the-art self-supervised methods by 4.7-11.0%/4.8-7.3% (PSNR/SSIM) on brain MRI data and by 43.6-50.5%/57.1-77.1% (PSNR/SSIM) on dual-energy CT X-ray microscopy data with respect to the noisy baseline. Our experiments on different real measured data sets indicate that Noise2Contrast training generalizes to other multi-contrast imaging modalities.
Gradient-Based Geometry Learning for Fan-Beam CT Reconstruction
Dec 05, 2022Abstract:Incorporating computed tomography (CT) reconstruction operators into differentiable pipelines has proven beneficial in many applications. Such approaches usually focus on the projection data and keep the acquisition geometry fixed. However, precise knowledge of the acquisition geometry is essential for high quality reconstruction results. In this paper, the differentiable formulation of fan-beam CT reconstruction is extended to the acquisition geometry. This allows to propagate gradient information from a loss function on the reconstructed image into the geometry parameters. As a proof-of-concept experiment, this idea is applied to rigid motion compensation. The cost function is parameterized by a trained neural network which regresses an image quality metric from the motion affected reconstruction alone. Using the proposed method, we are the first to optimize such an autofocus-inspired algorithm based on analytical gradients. The algorithm achieves a reduction in MSE by 35.5 % and an improvement in SSIM by 12.6 % over the motion affected reconstruction. Next to motion compensation, we see further use cases of our differentiable method for scanner calibration or hybrid techniques employing deep models.
On the Benefit of Dual-domain Denoising in a Self-supervised Low-dose CT Setting
Nov 03, 2022



Abstract:Computed tomography (CT) is routinely used for three-dimensional non-invasive imaging. Numerous data-driven image denoising algorithms were proposed to restore image quality in low-dose acquisitions. However, considerably less research investigates methods already intervening in the raw detector data due to limited access to suitable projection data or correct reconstruction algorithms. In this work, we present an end-to-end trainable CT reconstruction pipeline that contains denoising operators in both the projection and the image domain and that are optimized simultaneously without requiring ground-truth high-dose CT data. Our experiments demonstrate that including an additional projection denoising operator improved the overall denoising performance by 82.4-94.1%/12.5-41.7% (PSNR/SSIM) on abdomen CT and 1.5-2.9%/0.4-0.5% (PSNR/SSIM) on XRM data relative to the low-dose baseline. We make our entire helical CT reconstruction framework publicly available that contains a raw projection rebinning step to render helical projection data suitable for differentiable fan-beam reconstruction operators and end-to-end learning.
Trainable Joint Bilateral Filters for Enhanced Prediction Stability in Low-dose CT
Jul 15, 2022
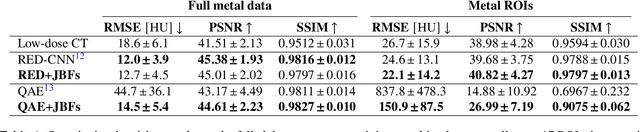
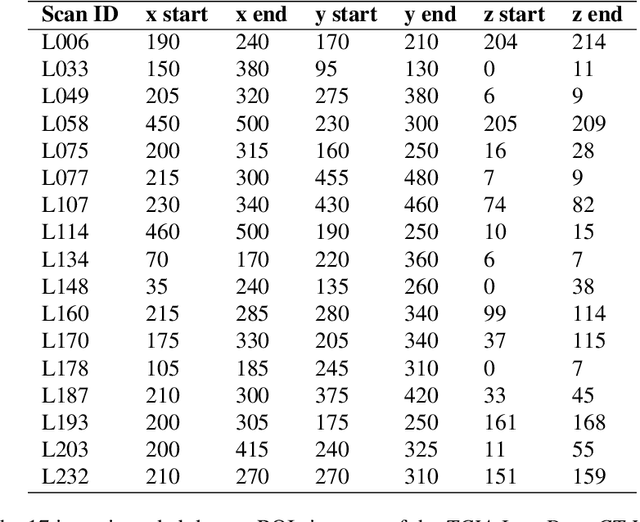
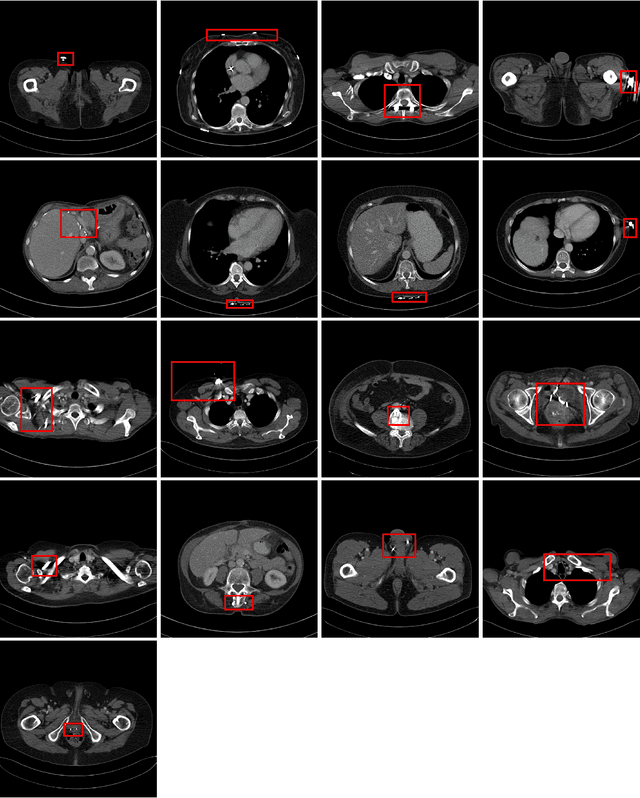
Abstract:Low-dose computed tomography (CT) denoising algorithms aim to enable reduced patient dose in routine CT acquisitions while maintaining high image quality. Recently, deep learning~(DL)-based methods were introduced, outperforming conventional denoising algorithms on this task due to their high model capacity. However, for the transition of DL-based denoising to clinical practice, these data-driven approaches must generalize robustly beyond the seen training data. We, therefore, propose a hybrid denoising approach consisting of a set of trainable joint bilateral filters (JBFs) combined with a convolutional DL-based denoising network to predict the guidance image. Our proposed denoising pipeline combines the high model capacity enabled by DL-based feature extraction with the reliability of the conventional JBF. The pipeline's ability to generalize is demonstrated by training on abdomen CT scans without metal implants and testing on abdomen scans with metal implants as well as on head CT data. When embedding two well-established DL-based denoisers (RED-CNN/QAE) in our pipeline, the denoising performance is improved by $10\,\%$/$82\,\%$ (RMSE) and $3\,\%$/$81\,\%$ (PSNR) in regions containing metal and by $6\,\%$/$78\,\%$ (RMSE) and $2\,\%$/$4\,\%$ (PSNR) on head CT data, compared to the respective vanilla model. Concluding, the proposed trainable JBFs limit the error bound of deep neural networks to facilitate the applicability of DL-based denoisers in low-dose CT pipelines.
ConFUDA: Contrastive Fewshot Unsupervised Domain Adaptation for Medical Image Segmentation
Jun 08, 2022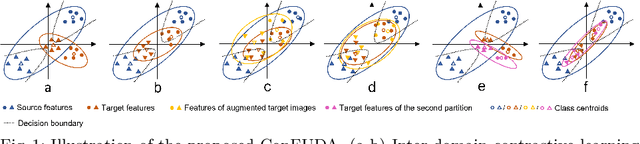
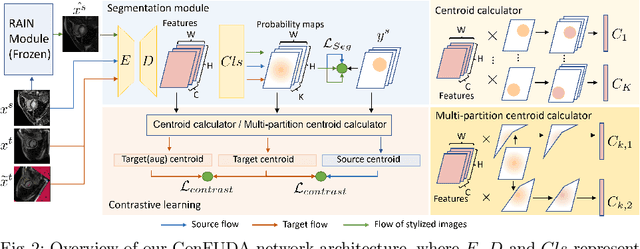
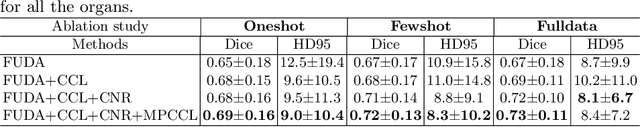
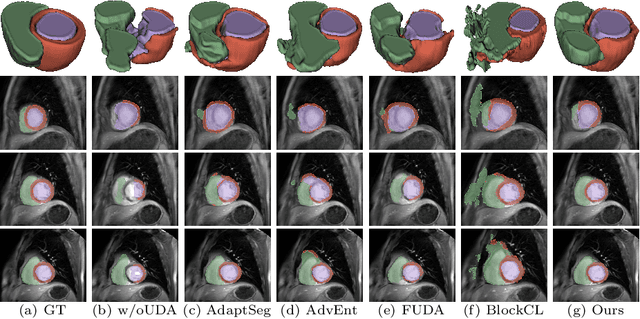
Abstract:Unsupervised domain adaptation (UDA) aims to transfer knowledge learned from a labeled source domain to an unlabeled target domain. Contrastive learning (CL) in the context of UDA can help to better separate classes in feature space. However, in image segmentation, the large memory footprint due to the computation of the pixel-wise contrastive loss makes it prohibitive to use. Furthermore, labeled target data is not easily available in medical imaging, and obtaining new samples is not economical. As a result, in this work, we tackle a more challenging UDA task when there are only a few (fewshot) or a single (oneshot) image available from the target domain. We apply a style transfer module to mitigate the scarcity of target samples. Then, to align the source and target features and tackle the memory issue of the traditional contrastive loss, we propose the centroid-based contrastive learning (CCL) and a centroid norm regularizer (CNR) to optimize the contrastive pairs in both direction and magnitude. In addition, we propose multi-partition centroid contrastive learning (MPCCL) to further reduce the variance in the target features. Fewshot evaluation on MS-CMRSeg dataset demonstrates that ConFUDA improves the segmentation performance by 0.34 of the Dice score on the target domain compared with the baseline, and 0.31 Dice score improvement in a more rigorous oneshot setting.
Few-shot Unsupervised Domain Adaptation for Multi-modal Cardiac Image Segmentation
Jan 28, 2022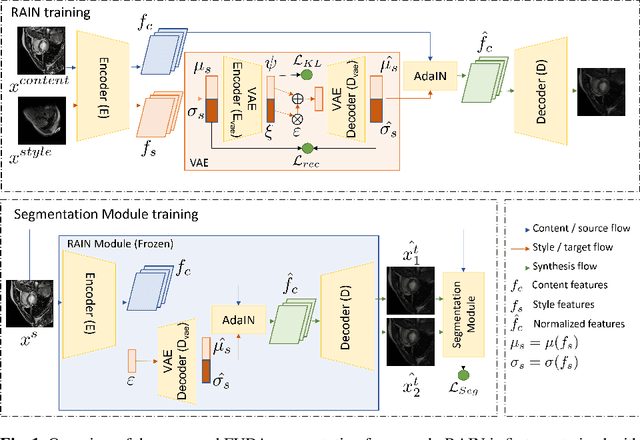
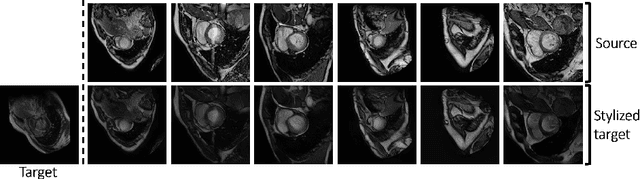
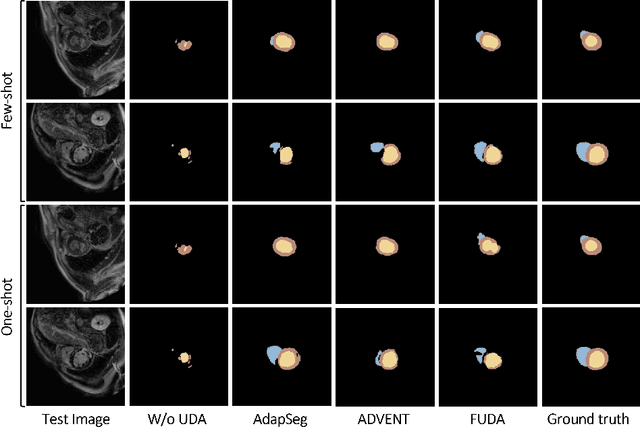
Abstract:Unsupervised domain adaptation (UDA) methods intend to reduce the gap between source and target domains by using unlabeled target domain and labeled source domain data, however, in the medical domain, target domain data may not always be easily available, and acquiring new samples is generally time-consuming. This restricts the development of UDA methods for new domains. In this paper, we explore the potential of UDA in a more challenging while realistic scenario where only one unlabeled target patient sample is available. We call it Few-shot Unsupervised Domain adaptation (FUDA). We first generate target-style images from source images and explore diverse target styles from a single target patient with Random Adaptive Instance Normalization (RAIN). Then, a segmentation network is trained in a supervised manner with the generated target images. Our experiments demonstrate that FUDA improves the segmentation performance by 0.33 of Dice score on the target domain compared with the baseline, and it also gives 0.28 of Dice score improvement in a more rigorous one-shot setting. Our code is available at \url{https://github.com/MingxuanGu/Few-shot-UDA}.
Ultra Low-Parameter Denoising: Trainable Bilateral Filter Layers in Computed Tomography
Jan 25, 2022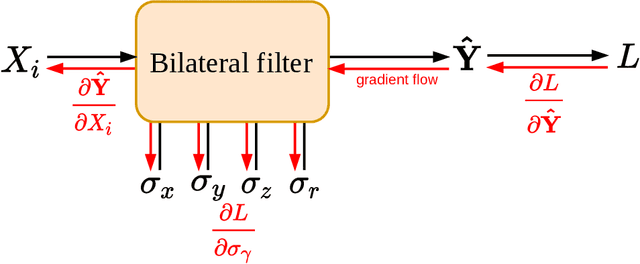
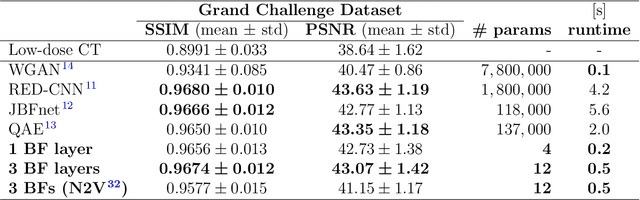
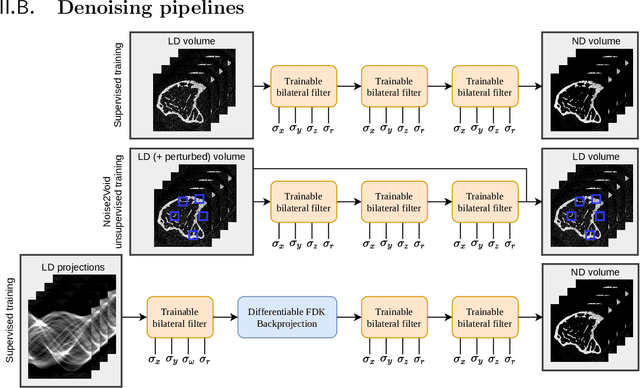
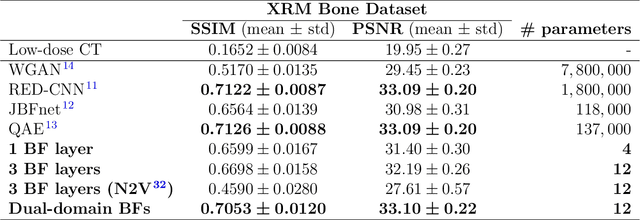
Abstract:Computed tomography is widely used as an imaging tool to visualize three-dimensional structures with expressive bone-soft tissue contrast. However, CT resolution and radiation dose are tightly entangled, highlighting the importance of low-dose CT combined with sophisticated denoising algorithms. Most data-driven denoising techniques are based on deep neural networks and, therefore, contain hundreds of thousands of trainable parameters, making them incomprehensible and prone to prediction failures. Developing understandable and robust denoising algorithms achieving state-of-the-art performance helps to minimize radiation dose while maintaining data integrity. This work presents an open-source CT denoising framework based on the idea of bilateral filtering. We propose a bilateral filter that can be incorporated into a deep learning pipeline and optimized in a purely data-driven way by calculating the gradient flow toward its hyperparameters and its input. Denoising in pure image-to-image pipelines and across different domains such as raw detector data and reconstructed volume, using a differentiable backprojection layer, is demonstrated. Although only using three spatial parameters and one range parameter per filter layer, the proposed denoising pipelines can compete with deep state-of-the-art denoising architectures with several hundred thousand parameters. Competitive denoising performance is achieved on x-ray microscope bone data (0.7053 and 33.10) and the 2016 Low Dose CT Grand Challenge dataset (0.9674 and 43.07) in terms of SSIM and PSNR. Due to the extremely low number of trainable parameters with well-defined effect, prediction reliance and data integrity is guaranteed at any time in the proposed pipelines, in contrast to most other deep learning-based denoising architectures.
Learned Cone-Beam CT Reconstruction Using Neural Ordinary Differential Equations
Jan 19, 2022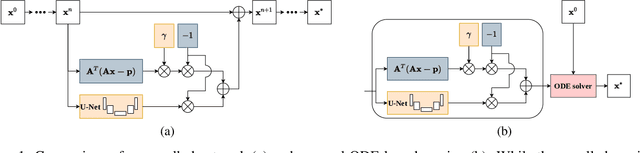
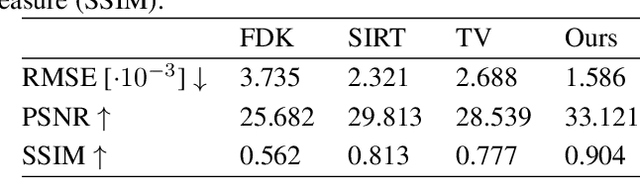
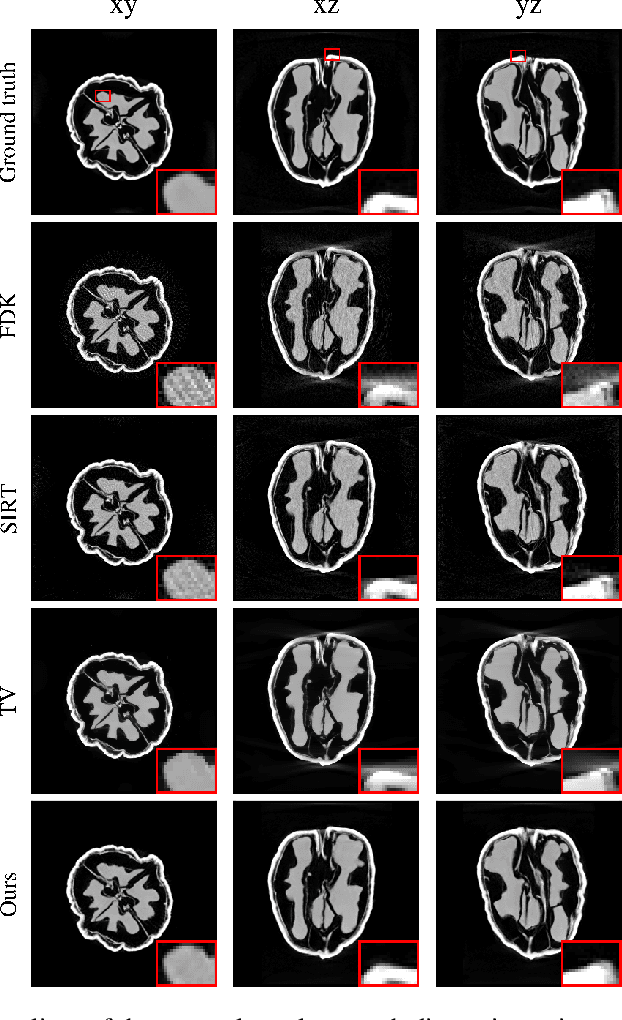
Abstract:Learned iterative reconstruction algorithms for inverse problems offer the flexibility to combine analytical knowledge about the problem with modules learned from data. This way, they achieve high reconstruction performance while ensuring consistency with the measured data. In computed tomography, extending such approaches from 2D fan-beam to 3D cone-beam data is challenging due to the prohibitively high GPU memory that would be needed to train such models. This paper proposes to use neural ordinary differential equations to solve the reconstruction problem in a residual formulation via numerical integration. For training, there is no need to backpropagate through several unrolled network blocks nor through the internals of the solver. Instead, the gradients are obtained very memory-efficiently in the neural ODE setting allowing for training on a single consumer graphics card. The method is able to reduce the root mean squared error by over 30% compared to the best performing classical iterative reconstruction algorithm and produces high quality cone-beam reconstructions even in a sparse view scenario.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge