MingDe Lin
Analysis of the MICCAI Brain Tumor Segmentation -- Metastases (BraTS-METS) 2025 Lighthouse Challenge: Brain Metastasis Segmentation on Pre- and Post-treatment MRI
Apr 16, 2025Abstract:Despite continuous advancements in cancer treatment, brain metastatic disease remains a significant complication of primary cancer and is associated with an unfavorable prognosis. One approach for improving diagnosis, management, and outcomes is to implement algorithms based on artificial intelligence for the automated segmentation of both pre- and post-treatment MRI brain images. Such algorithms rely on volumetric criteria for lesion identification and treatment response assessment, which are still not available in clinical practice. Therefore, it is critical to establish tools for rapid volumetric segmentations methods that can be translated to clinical practice and that are trained on high quality annotated data. The BraTS-METS 2025 Lighthouse Challenge aims to address this critical need by establishing inter-rater and intra-rater variability in dataset annotation by generating high quality annotated datasets from four individual instances of segmentation by neuroradiologists while being recorded on video (two instances doing "from scratch" and two instances after AI pre-segmentation). This high-quality annotated dataset will be used for testing phase in 2025 Lighthouse challenge and will be publicly released at the completion of the challenge. The 2025 Lighthouse challenge will also release the 2023 and 2024 segmented datasets that were annotated using an established pipeline of pre-segmentation, student annotation, two neuroradiologists checking, and one neuroradiologist finalizing the process. It builds upon its previous edition by including post-treatment cases in the dataset. Using these high-quality annotated datasets, the 2025 Lighthouse challenge plans to test benchmark algorithms for automated segmentation of pre-and post-treatment brain metastases (BM), trained on diverse and multi-institutional datasets of MRI images obtained from patients with brain metastases.
The Brain Tumor Segmentation (BraTS-METS) Challenge 2023: Brain Metastasis Segmentation on Pre-treatment MRI
Jun 01, 2023



Abstract:Clinical monitoring of metastatic disease to the brain can be a laborious and time-consuming process, especially in cases involving multiple metastases when the assessment is performed manually. The Response Assessment in Neuro-Oncology Brain Metastases (RANO-BM) guideline, which utilizes the unidimensional longest diameter, is commonly used in clinical and research settings to evaluate response to therapy in patients with brain metastases. However, accurate volumetric assessment of the lesion and surrounding peri-lesional edema holds significant importance in clinical decision-making and can greatly enhance outcome prediction. The unique challenge in performing segmentations of brain metastases lies in their common occurrence as small lesions. Detection and segmentation of lesions that are smaller than 10 mm in size has not demonstrated high accuracy in prior publications. The brain metastases challenge sets itself apart from previously conducted MICCAI challenges on glioma segmentation due to the significant variability in lesion size. Unlike gliomas, which tend to be larger on presentation scans, brain metastases exhibit a wide range of sizes and tend to include small lesions. We hope that the BraTS-METS dataset and challenge will advance the field of automated brain metastasis detection and segmentation.
Hepatocellular Carcinoma Intra-arterial Treatment Response Prediction for Improved Therapeutic Decision-Making
Dec 01, 2019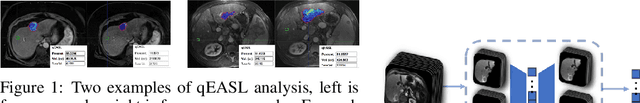
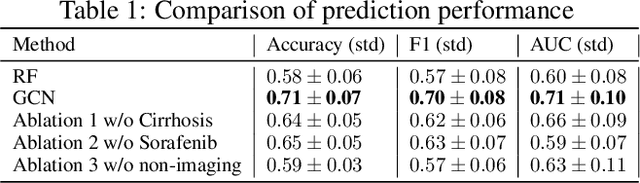
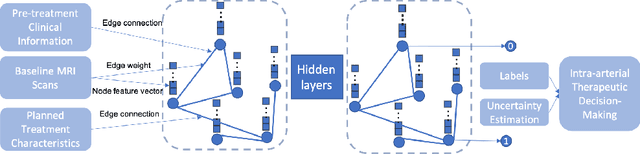
Abstract:This work proposes a pipeline to predict treatment response to intra-arterial therapy of patients with Hepatocellular Carcinoma (HCC) for improved therapeutic decision-making. Our graph neural network model seamlessly combines heterogeneous inputs of baseline MR scans, pre-treatment clinical information, and planned treatment characteristics and has been validated on patients with HCC treated by transarterial chemoembolization (TACE). It achieves Accuracy of $0.713 \pm 0.075$, F1 of $0.702 \pm 0.082$ and AUC of $0.710 \pm 0.108$. In addition, the pipeline incorporates uncertainty estimation to select hard cases and most align with the misclassified cases. The proposed pipeline arrives at more informed intra-arterial therapeutic decisions for patients with HCC via improving model accuracy and incorporating uncertainty estimation.
Unsupervised Domain Adaptation via Disentangled Representations: Application to Cross-Modality Liver Segmentation
Aug 29, 2019
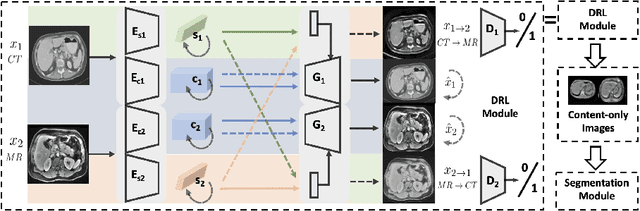
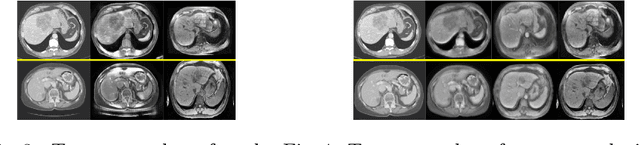
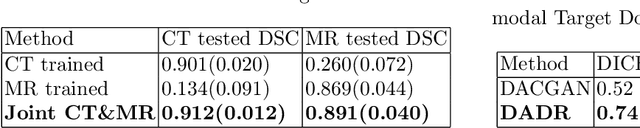
Abstract:A deep learning model trained on some labeled data from a certain source domain generally performs poorly on data from different target domains due to domain shifts. Unsupervised domain adaptation methods address this problem by alleviating the domain shift between the labeled source data and the unlabeled target data. In this work, we achieve cross-modality domain adaptation, i.e. between CT and MRI images, via disentangled representations. Compared to learning a one-to-one mapping as the state-of-art CycleGAN, our model recovers a many-to-many mapping between domains to capture the complex cross-domain relations. It preserves semantic feature-level information by finding a shared content space instead of a direct pixelwise style transfer. Domain adaptation is achieved in two steps. First, images from each domain are embedded into two spaces, a shared domain-invariant content space and a domain-specific style space. Next, the representation in the content space is extracted to perform a task. We validated our method on a cross-modality liver segmentation task, to train a liver segmentation model on CT images that also performs well on MRI. Our method achieved Dice Similarity Coefficient (DSC) of 0.81, outperforming a CycleGAN-based method of 0.72. Moreover, our model achieved good generalization to joint-domain learning, in which unpaired data from different modalities are jointly learned to improve the segmentation performance on each individual modality. Lastly, under a multi-modal target domain with significant diversity, our approach exhibited the potential for diverse image generation and remained effective with DSC of 0.74 on multi-phasic MRI while the CycleGAN-based method performed poorly with a DSC of only 0.52.
Domain-Agnostic Learning with Anatomy-Consistent Embedding for Cross-Modality Liver Segmentation
Aug 27, 2019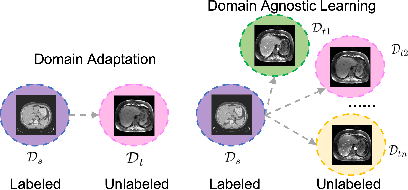
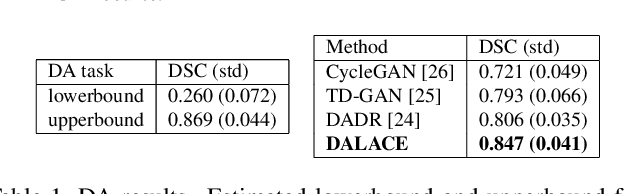
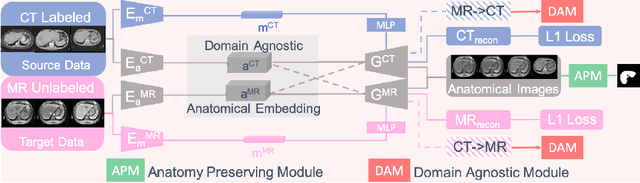
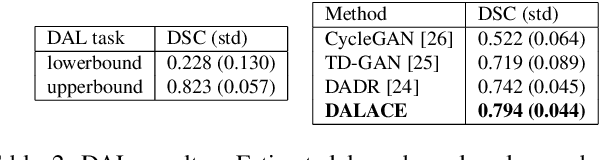
Abstract:Domain Adaptation (DA) has the potential to greatly help the generalization of deep learning models. However, the current literature usually assumes to transfer the knowledge from the source domain to a specific known target domain. Domain Agnostic Learning (DAL) proposes a new task of transferring knowledge from the source domain to data from multiple heterogeneous target domains. In this work, we propose the Domain-Agnostic Learning framework with Anatomy-Consistent Embedding (DALACE) that works on both domain-transfer and task-transfer to learn a disentangled representation, aiming to not only be invariant to different modalities but also preserve anatomical structures for the DA and DAL tasks in cross-modality liver segmentation. We validated and compared our model with state-of-the-art methods, including CycleGAN, Task Driven Generative Adversarial Network (TD-GAN), and Domain Adaptation via Disentangled Representations (DADR). For the DA task, our DALACE model outperformed CycleGAN, TD-GAN ,and DADR with DSC of 0.847 compared to 0.721, 0.793 and 0.806. For the DAL task, our model improved the performance with DSC of 0.794 from 0.522, 0.719 and 0.742 by CycleGAN, TD-GAN, and DADR. Further, we visualized the success of disentanglement, which added human interpretability of the learned meaningful representations. Through ablation analysis, we specifically showed the concrete benefits of disentanglement for downstream tasks and the role of supervision for better disentangled representation with segmentation consistency to be invariant to domains with the proposed Domain-Agnostic Module (DAM) and to preserve anatomical information with the proposed Anatomy-Preserving Module (APM).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge