Melissa Estevez
Ensuring Reliability of Curated EHR-Derived Data: The Validation of Accuracy for LLM/ML-Extracted Information and Data (VALID) Framework
Jun 09, 2025Abstract:Large language models (LLMs) are increasingly used to extract clinical data from electronic health records (EHRs), offering significant improvements in scalability and efficiency for real-world data (RWD) curation in oncology. However, the adoption of LLMs introduces new challenges in ensuring the reliability, accuracy, and fairness of extracted data, which are essential for research, regulatory, and clinical applications. Existing quality assurance frameworks for RWD and artificial intelligence do not fully address the unique error modes and complexities associated with LLM-extracted data. In this paper, we propose a comprehensive framework for evaluating the quality of clinical data extracted by LLMs. The framework integrates variable-level performance benchmarking against expert human abstraction, automated verification checks for internal consistency and plausibility, and replication analyses comparing LLM-extracted data to human-abstracted datasets or external standards. This multidimensional approach enables the identification of variables most in need of improvement, systematic detection of latent errors, and confirmation of dataset fitness-for-purpose in real-world research. Additionally, the framework supports bias assessment by stratifying metrics across demographic subgroups. By providing a rigorous and transparent method for assessing LLM-extracted RWD, this framework advances industry standards and supports the trustworthy use of AI-powered evidence generation in oncology research and practice.
Model-assisted cohort selection with bias analysis for generating large-scale cohorts from the EHR for oncology research
Jan 13, 2020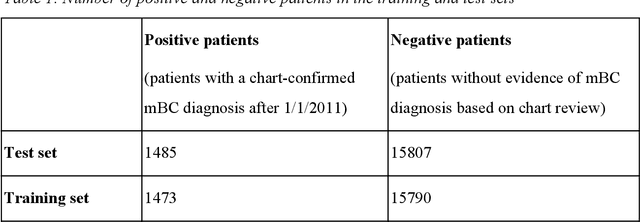
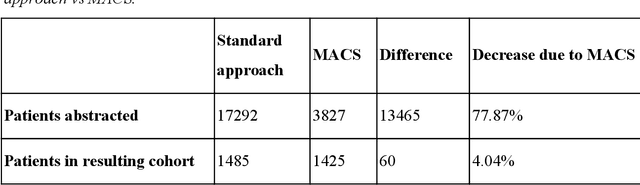
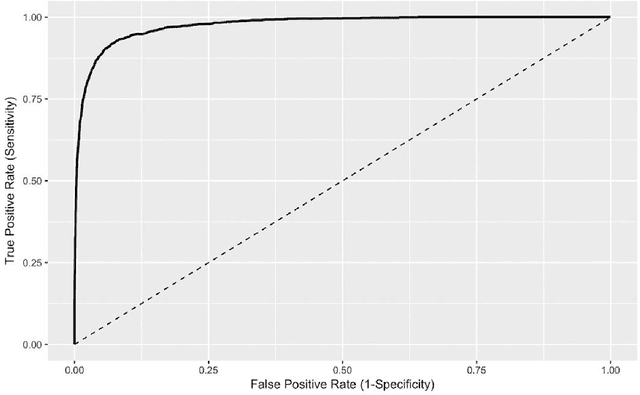
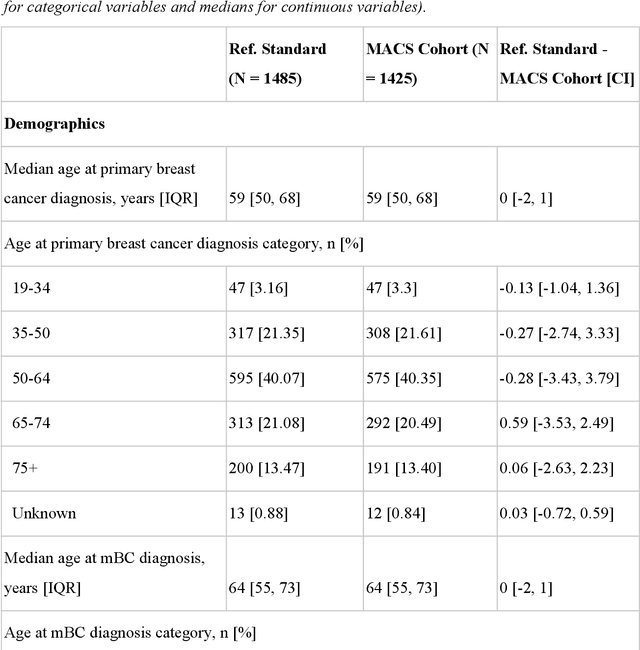
Abstract:Objective Electronic health records (EHRs) are a promising source of data for health outcomes research in oncology. A challenge in using EHR data is that selecting cohorts of patients often requires information in unstructured parts of the record. Machine learning has been used to address this, but even high-performing algorithms may select patients in a non-random manner and bias the resulting cohort. To improve the efficiency of cohort selection while measuring potential bias, we introduce a technique called Model-Assisted Cohort Selection (MACS) with Bias Analysis and apply it to the selection of metastatic breast cancer (mBC) patients. Materials and Methods We trained a model on 17,263 patients using term-frequency inverse-document-frequency (TF-IDF) and logistic regression. We used a test set of 17,292 patients to measure algorithm performance and perform Bias Analysis. We compared the cohort generated by MACS to the cohort that would have been generated without MACS as reference standard, first by comparing distributions of an extensive set of clinical and demographic variables and then by comparing the results of two analyses addressing existing example research questions. Results Our algorithm had an area under the curve (AUC) of 0.976, a sensitivity of 96.0%, and an abstraction efficiency gain of 77.9%. During Bias Analysis, we found no large differences in baseline characteristics and no differences in the example analyses. Conclusion MACS with bias analysis can significantly improve the efficiency of cohort selection on EHR data while instilling confidence that outcomes research performed on the resulting cohort will not be biased.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge