Md. Kamrul Hasan
Systems and Software Lab, Department of Computer Science and Engineering, Islamic University of Technology
FUGC: Benchmarking Semi-Supervised Learning Methods for Cervical Segmentation
Jan 22, 2026Abstract:Accurate segmentation of cervical structures in transvaginal ultrasound (TVS) is critical for assessing the risk of spontaneous preterm birth (PTB), yet the scarcity of labeled data limits the performance of supervised learning approaches. This paper introduces the Fetal Ultrasound Grand Challenge (FUGC), the first benchmark for semi-supervised learning in cervical segmentation, hosted at ISBI 2025. FUGC provides a dataset of 890 TVS images, including 500 training images, 90 validation images, and 300 test images. Methods were evaluated using the Dice Similarity Coefficient (DSC), Hausdorff Distance (HD), and runtime (RT), with a weighted combination of 0.4/0.4/0.2. The challenge attracted 10 teams with 82 participants submitting innovative solutions. The best-performing methods for each individual metric achieved 90.26\% mDSC, 38.88 mHD, and 32.85 ms RT, respectively. FUGC establishes a standardized benchmark for cervical segmentation, demonstrates the efficacy of semi-supervised methods with limited labeled data, and provides a foundation for AI-assisted clinical PTB risk assessment.
Multi-Stage Residual-Aware Unsupervised Deep Learning Framework for Consistent Ultrasound Strain Elastography
Nov 19, 2025Abstract:Ultrasound Strain Elastography (USE) is a powerful non-invasive imaging technique for assessing tissue mechanical properties, offering crucial diagnostic value across diverse clinical applications. However, its clinical application remains limited by tissue decorrelation noise, scarcity of ground truth, and inconsistent strain estimation under different deformation conditions. Overcoming these barriers, we propose MUSSE-Net, a residual-aware, multi-stage unsupervised sequential deep learning framework designed for robust and consistent strain estimation. At its backbone lies our proposed USSE-Net, an end-to-end multi-stream encoder-decoder architecture that parallelly processes pre- and post-deformation RF sequences to estimate displacement fields and axial strains. The novel architecture incorporates Context-Aware Complementary Feature Fusion (CACFF)-based encoder with Tri-Cross Attention (TCA) bottleneck with a Cross-Attentive Fusion (CAF)-based sequential decoder. To ensure temporal coherence and strain stability across varying deformation levels, this architecture leverages a tailored consistency loss. Finally, with the MUSSE-Net framework, a secondary residual refinement stage further enhances accuracy and suppresses noise. Extensive validation on simulation, in vivo, and private clinical datasets from Bangladesh University of Engineering and Technology (BUET) medical center, demonstrates MUSSE-Net's outperformed existing unsupervised approaches. On MUSSE-Net achieves state-of-the-art performance with a target SNR of 24.54, background SNR of 132.76, CNR of 59.81, and elastographic SNR of 9.73 on simulation data. In particular, on the BUET dataset, MUSSE-Net produces strain maps with enhanced lesion-to-background contrast and significant noise suppression yielding clinically interpretable strain patterns.
EXGnet: a single-lead explainable-AI guided multiresolution network with train-only quantitative features for trustworthy ECG arrhythmia classification
Jun 14, 2025



Abstract:Background: Deep learning has significantly advanced ECG arrhythmia classification, enabling high accuracy in detecting various cardiac conditions. The use of single-lead ECG systems is crucial for portable devices, as they offer convenience and accessibility for continuous monitoring in diverse settings. However, the interpretability and reliability of deep learning models in clinical applications poses challenges due to their black-box nature. Methods: To address these challenges, we propose EXGnet, a single-lead, trustworthy ECG arrhythmia classification network that integrates multiresolution feature extraction with Explainable Artificial Intelligence (XAI) guidance and train only quantitative features. Results: Trained on two public datasets, including Chapman and Ningbo, EXGnet demonstrates superior performance through key metrics such as Accuracy, F1-score, Sensitivity, and Specificity. The proposed method achieved average five fold accuracy of 98.762%, and 96.932% and average F1-score of 97.910%, and 95.527% on the Chapman and Ningbo datasets, respectively. Conclusions: By employing XAI techniques, specifically Grad-CAM, the model provides visual insights into the relevant ECG segments it analyzes, thereby enhancing clinician trust in its predictions. While quantitative features further improve classification performance, they are not required during testing, making the model suitable for real-world applications. Overall, EXGnet not only achieves better classification accuracy but also addresses the critical need for interpretability in deep learning, facilitating broader adoption in portable ECG monitoring.
SW-ViT: A Spatio-Temporal Vision Transformer Network with Post Denoiser for Sequential Multi-Push Ultrasound Shear Wave Elastography
May 24, 2025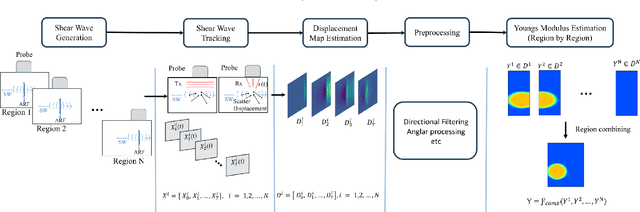



Abstract:Objective: Ultrasound Shear Wave Elastography (SWE) demonstrates great potential in assessing soft-tissue pathology by mapping tissue stiffness, which is linked to malignancy. Traditional SWE methods have shown promise in estimating tissue elasticity, yet their susceptibility to noise interference, reliance on limited training data, and inability to generate segmentation masks concurrently present notable challenges to accuracy and reliability. Approach: In this paper, we propose SW-ViT, a novel two-stage deep learning framework for SWE that integrates a CNN-Spatio-Temporal Vision Transformer-based reconstruction network with an efficient Transformer-based post-denoising network. The first stage uses a 3D ResNet encoder with multi-resolution spatio-temporal Transformer blocks that capture spatial and temporal features, followed by a squeeze-and-excitation attention decoder that reconstructs 2D stiffness maps. To address data limitations, a patch-based training strategy is adopted for localized learning and reconstruction. In the second stage, a denoising network with a shared encoder and dual decoders processes inclusion and background regions to produce a refined stiffness map and segmentation mask. A hybrid loss combining regional, smoothness, fusion, and Intersection over Union (IoU) components ensures improvements in both reconstruction and segmentation. Results: On simulated data, our method achieves PSNR of 32.68 dB, CNR of 46.78 dB, and SSIM of 0.995. On phantom data, results include PSNR of 21.11 dB, CNR of 42.14 dB, and SSIM of 0.936. Segmentation IoU values reach 0.949 (simulation) and 0.738 (phantom) with ASSD values being 0.184 and 1.011, respectively. Significance: SW-ViT delivers robust, high-quality elasticity map estimates from noisy SWE data and holds clear promise for clinical application.
Motion-enhancement to Echocardiography Segmentation via Inserting a Temporal Attention Module: An Efficient, Adaptable, and Scalable Approach
Jan 24, 2025



Abstract:Cardiac anatomy segmentation is essential for clinical assessment of cardiac function and disease diagnosis to inform treatment and intervention. In performing segmentation, deep learning (DL) algorithms improved accuracy significantly compared to traditional image processing approaches. More recently, studies showed that enhancing DL segmentation with motion information can further improve it. A range of methods for injecting motion information has been proposed, but many of them increase the dimensionality of input images (which is computationally expensive) or have not used an optimal method to insert motion information, such as non-DL registration, non-attention-based networks or single-headed attention. Here, we present a novel, computation-efficient alternative where a novel, scalable temporal attention module (TAM) extracts temporal feature interactions multiple times and where TAM has a multi-headed, KQV projection cross-attention architecture. The module can be seamlessly integrated into a wide range of existing CNN- or Transformer-based networks, providing novel flexibility for inclusion in future implementations. Extensive evaluations on different cardiac datasets, 2D echocardiography (CAMUS), and 3D echocardiography (MITEA) demonstrate the model's effectiveness when integrated into well-established backbone networks like UNet, FCN8s, UNetR, SwinUNetR, and the recent I2UNet. We further find that the optimized TAM-enhanced FCN8s network performs well compared to contemporary alternatives. Our results confirm TAM's robustness, scalability, and generalizability across diverse datasets and backbones.
A Constrained Optimization approach for Ultrasound Shear Wave Speed Estimation with Time-Lateral Plane Cleaning
Jul 30, 2024



Abstract:Ultrasound shear wave elastography (SWE) is a noninvasive way to measure stiffness of soft tissue for medical diagnosis. In SWE imaging, an acoustic radiation force induces tissue displacement, which creates shear waves (SWs) that travel laterally through the medium. Finding the lateral arrival times of SWs at different tissue locations helps figure out the shear wave speed (SWS), which is directly linked to the stiffness of the medium. Traditional SWS estimation techniques, however, are not noise resilient enough handling noise and reflection artifacts filled data. This paper proposes new techniques to estimate SWS in both time and frequency domains. These new methods optimize a loss function that is based on the lateral signal shift parameter between known locations and is constrained by neighborhood displacement group shift determined from the time-lateral plane-denoised SW propagation data. The proposed constrained optimization is formed by coupling losses of local particles with a Gaussian kernel giving an optimum arrival time for the center particle by enforcing stiffness homogeneity in a small neighborhood to enable inherent noise resilience. The denoising scheme involves isolating the transitioning SW profile in each time-lateral plane, creating a parameterized mask. Moreover, lateral interpolation is performed to enhance reconstruction resolution and obtain increased displacement groups to enhance the reliability of the optimization. The proposed noise robust scheme is tested on a simulation and three experimental datasets. The performance of the method is compared with 3 ToF and 2 frequency-domain methods. The evaluations show visually and quantitatively superior and noise-robust reconstructions compared to state-of-the-art methods. Due to its high contrast and minimal error, the proposed technique can find its application in tissue health inspection and cancer diagnosis.
Robust CNN Multi-Nested-LSTM Framework with Compound Loss for Patch-based Multi-Push Ultrasound Shear Wave Imaging and Segmentation
Jul 30, 2024



Abstract:Ultrasound Shear Wave Elastography (SWE) is a noteworthy tool for in-vivo noninvasive tissue pathology assessment. State-of-the-art techniques can generate reasonable estimates of tissue elasticity, but high-quality and noise-resiliency in SWE reconstruction have yet to demonstrate advancements. In this work, we propose a two-stage DL pipeline producing reliable reconstructions and denoise said reconstructions to obtain lower noise prevailing elasticity mappings. The reconstruction network consists of a Resnet3D Encoder to extract temporal context from the sequential multi-push data. The encoded features are sent to multiple Nested CNN LSTMs which process them in a temporal attention-guided windowing basis and map the 3D features to 2D using FFT-attention, which are then decoded into an elasticity map as primary reconstruction. The 2D maps from each multi-push region are merged and cleaned using a dual-decoder denoiser network, which independently denoises foreground and background before fusion. The post-denoiser generates a higher-quality reconstruction and an inclusion-segmentation mask. A multi-objective loss is designed to accommodate the denoising, fusing, and segmentation processes. The method is validated on sequential multi-push SWE motion data with multiple overlapping regions. A patch-based training procedure is introduced with network modifications to handle data scarcity. Evaluations produce 32.66dB PSNR, 43.19dB CNR in noisy simulation, and 22.44dB PSNR, 36.88dB CNR in experimental data, across all test samples. Moreover, IoUs (0.909 and 0.781) were quite satisfactory in the datasets. After comparing with other reported deep-learning approaches, our method proves quantitatively and qualitatively superior in dealing with noise influences in SWE data. From a performance point of view, our deep-learning pipeline has the potential to become utilitarian in the clinical domain.
BdSLW60: A Word-Level Bangla Sign Language Dataset
Feb 13, 2024



Abstract:Sign language discourse is an essential mode of daily communication for the deaf and hard-of-hearing people. However, research on Bangla Sign Language (BdSL) faces notable limitations, primarily due to the lack of datasets. Recognizing wordlevel signs in BdSL (WL-BdSL) presents a multitude of challenges, including the need for well-annotated datasets, capturing the dynamic nature of sign gestures from facial or hand landmarks, developing suitable machine learning or deep learning-based models with substantial video samples, and so on. In this paper, we address these challenges by creating a comprehensive BdSL word-level dataset named BdSLW60 in an unconstrained and natural setting, allowing positional and temporal variations and allowing sign users to change hand dominance freely. The dataset encompasses 60 Bangla sign words, with a significant scale of 9307 video trials provided by 18 signers under the supervision of a sign language professional. The dataset was rigorously annotated and cross-checked by 60 annotators. We also introduced a unique approach of a relative quantization-based key frame encoding technique for landmark based sign gesture recognition. We report the benchmarking of our BdSLW60 dataset using the Support Vector Machine (SVM) with testing accuracy up to 67.6% and an attention-based bi-LSTM with testing accuracy up to 75.1%. The dataset is available at https://www.kaggle.com/datasets/hasaniut/bdslw60 and the code base is accessible from https://github.com/hasanssl/BdSLW60_Code.
Towards Automated Recipe Genre Classification using Semi-Supervised Learning
Oct 24, 2023



Abstract:Sharing cooking recipes is a great way to exchange culinary ideas and provide instructions for food preparation. However, categorizing raw recipes found online into appropriate food genres can be challenging due to a lack of adequate labeled data. In this study, we present a dataset named the ``Assorted, Archetypal, and Annotated Two Million Extended (3A2M+) Cooking Recipe Dataset" that contains two million culinary recipes labeled in respective categories with extended named entities extracted from recipe descriptions. This collection of data includes various features such as title, NER, directions, and extended NER, as well as nine different labels representing genres including bakery, drinks, non-veg, vegetables, fast food, cereals, meals, sides, and fusions. The proposed pipeline named 3A2M+ extends the size of the Named Entity Recognition (NER) list to address missing named entities like heat, time or process from the recipe directions using two NER extraction tools. 3A2M+ dataset provides a comprehensive solution to the various challenging recipe-related tasks, including classification, named entity recognition, and recipe generation. Furthermore, we have demonstrated traditional machine learning, deep learning and pre-trained language models to classify the recipes into their corresponding genre and achieved an overall accuracy of 98.6\%. Our investigation indicates that the title feature played a more significant role in classifying the genre.
Multi-scale, Data-driven and Anatomically Constrained Deep Learning Image Registration for Adult and Fetal Echocardiography
Sep 11, 2023



Abstract:Temporal echocardiography image registration is a basis for clinical quantifications such as cardiac motion estimation, myocardial strain assessments, and stroke volume quantifications. In past studies, deep learning image registration (DLIR) has shown promising results and is consistently accurate and precise, requiring less computational time. We propose that a greater focus on the warped moving image's anatomic plausibility and image quality can support robust DLIR performance. Further, past implementations have focused on adult echocardiography, and there is an absence of DLIR implementations for fetal echocardiography. We propose a framework that combines three strategies for DLIR in both fetal and adult echo: (1) an anatomic shape-encoded loss to preserve physiological myocardial and left ventricular anatomical topologies in warped images; (2) a data-driven loss that is trained adversarially to preserve good image texture features in warped images; and (3) a multi-scale training scheme of a data-driven and anatomically constrained algorithm to improve accuracy. Our tests show that good anatomical topology and image textures are strongly linked to shape-encoded and data-driven adversarial losses. They improve different aspects of registration performance in a non-overlapping way, justifying their combination. Despite fundamental distinctions between adult and fetal echo images, we show that these strategies can provide excellent registration results in both adult and fetal echocardiography using the publicly available CAMUS adult echo dataset and our private multi-demographic fetal echo dataset. Our approach outperforms traditional non-DL gold standard registration approaches, including Optical Flow and Elastix. Registration improvements could be translated to more accurate and precise clinical quantification of cardiac ejection fraction, demonstrating a potential for translation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge