Maxwell Reynolds
A Cautionary Tale of Self-Supervised Learning for Imaging Biomarkers: Alzheimer's Disease Case Study
Jan 23, 2026Abstract:Discovery of sensitive and biologically grounded biomarkers is essential for early detection and monitoring of Alzheimer's disease (AD). Structural MRI is widely available but typically relies on hand-crafted features such as cortical thickness or volume. We ask whether self-supervised learning (SSL) can uncover more powerful biomarkers from the same data. Existing SSL methods underperform FreeSurfer-derived features in disease classification, conversion prediction, and amyloid status prediction. We introduce Residual Noise Contrastive Estimation (R-NCE), a new SSL framework that integrates auxiliary FreeSurfer features while maximizing additional augmentation-invariant information. R-NCE outperforms traditional features and existing SSL methods across multiple benchmarks, including AD conversion prediction. To assess biological relevance, we derive Brain Age Gap (BAG) measures and perform genome-wide association studies. R-NCE-BAG shows high heritability and associations with MAPT and IRAG1, with enrichment in astrocytes and oligodendrocytes, indicating sensitivity to neurodegenerative and cerebrovascular processes.
Data Scaling Laws for Radiology Foundation Models
Sep 16, 2025Abstract:Foundation vision encoders such as CLIP and DINOv2, trained on web-scale data, exhibit strong transfer performance across tasks and datasets. However, medical imaging foundation models remain constrained by smaller datasets, limiting our understanding of how data scale and pretraining paradigms affect performance in this setting. In this work, we systematically study continual pretraining of two vision encoders, MedImageInsight (MI2) and RAD-DINO representing the two major encoder paradigms CLIP and DINOv2, on up to 3.5M chest x-rays from a single institution, holding compute and evaluation protocols constant. We evaluate on classification (radiology findings, lines and tubes), segmentation (lines and tubes), and radiology report generation. While prior work has primarily focused on tasks related to radiology findings, we include lines and tubes tasks to counterbalance this bias and evaluate a model's ability to extract features that preserve continuity along elongated structures. Our experiments show that MI2 scales more effectively for finding-related tasks, while RAD-DINO is stronger on tube-related tasks. Surprisingly, continually pretraining MI2 with both reports and structured labels using UniCL improves performance, underscoring the value of structured supervision at scale. We further show that for some tasks, as few as 30k in-domain samples are sufficient to surpass open-weights foundation models. These results highlight the utility of center-specific continual pretraining, enabling medical institutions to derive significant performance gains by utilizing in-domain data.
Adversarial Consistency for Single Domain Generalization in Medical Image Segmentation
Jun 29, 2022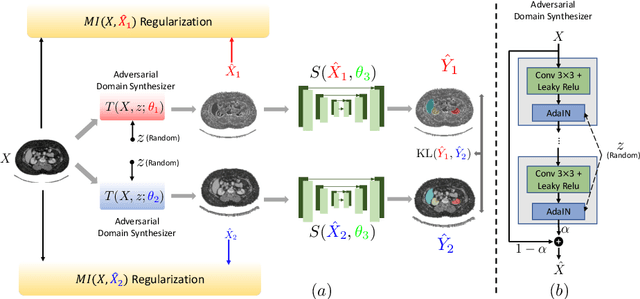
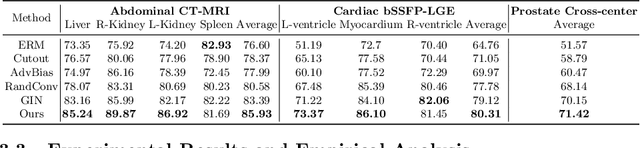
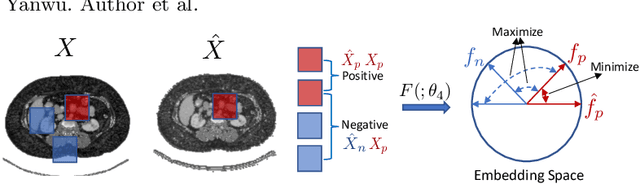
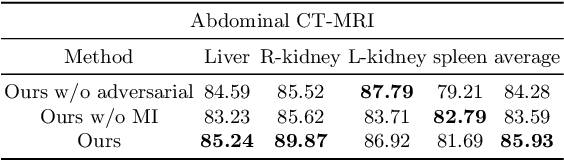
Abstract:An organ segmentation method that can generalize to unseen contrasts and scanner settings can significantly reduce the need for retraining of deep learning models. Domain Generalization (DG) aims to achieve this goal. However, most DG methods for segmentation require training data from multiple domains during training. We propose a novel adversarial domain generalization method for organ segmentation trained on data from a \emph{single} domain. We synthesize the new domains via learning an adversarial domain synthesizer (ADS) and presume that the synthetic domains cover a large enough area of plausible distributions so that unseen domains can be interpolated from synthetic domains. We propose a mutual information regularizer to enforce the semantic consistency between images from the synthetic domains, which can be estimated by patch-level contrastive learning. We evaluate our method for various organ segmentation for unseen modalities, scanning protocols, and scanner sites.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge