Matthew Ragoza
University of Pittsburgh
Adversarial Consistency for Single Domain Generalization in Medical Image Segmentation
Jun 29, 2022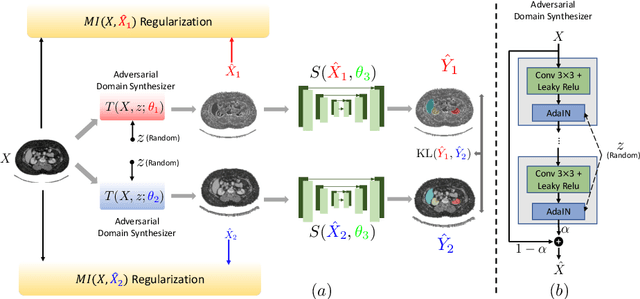
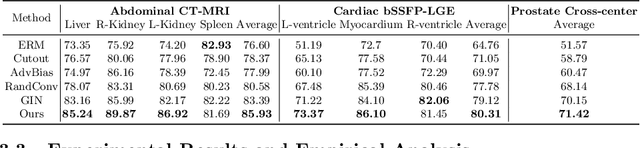
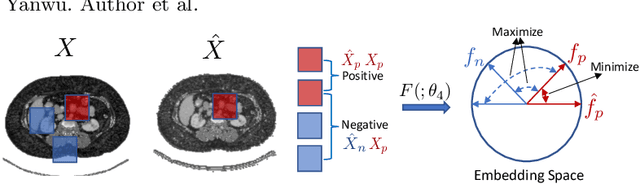
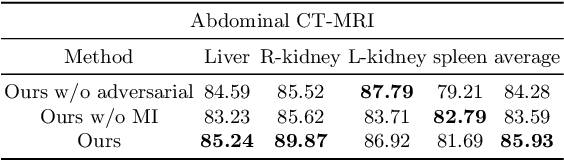
Abstract:An organ segmentation method that can generalize to unseen contrasts and scanner settings can significantly reduce the need for retraining of deep learning models. Domain Generalization (DG) aims to achieve this goal. However, most DG methods for segmentation require training data from multiple domains during training. We propose a novel adversarial domain generalization method for organ segmentation trained on data from a \emph{single} domain. We synthesize the new domains via learning an adversarial domain synthesizer (ADS) and presume that the synthetic domains cover a large enough area of plausible distributions so that unseen domains can be interpolated from synthetic domains. We propose a mutual information regularizer to enforce the semantic consistency between images from the synthetic domains, which can be estimated by patch-level contrastive learning. We evaluate our method for various organ segmentation for unseen modalities, scanning protocols, and scanner sites.
Generating 3D Molecules Conditional on Receptor Binding Sites with Deep Generative Models
Oct 28, 2021
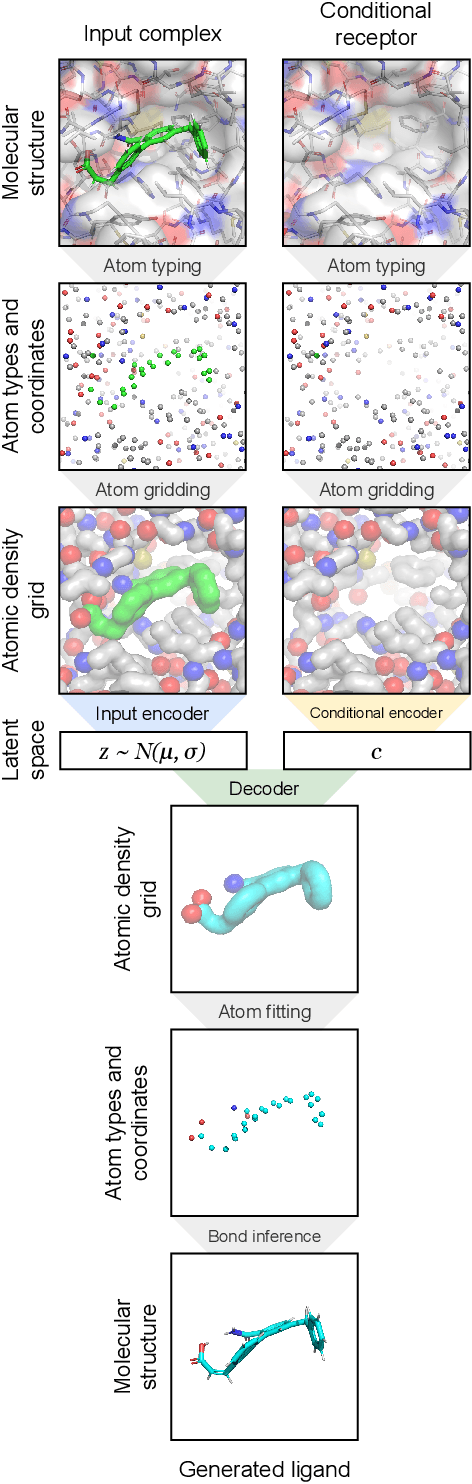
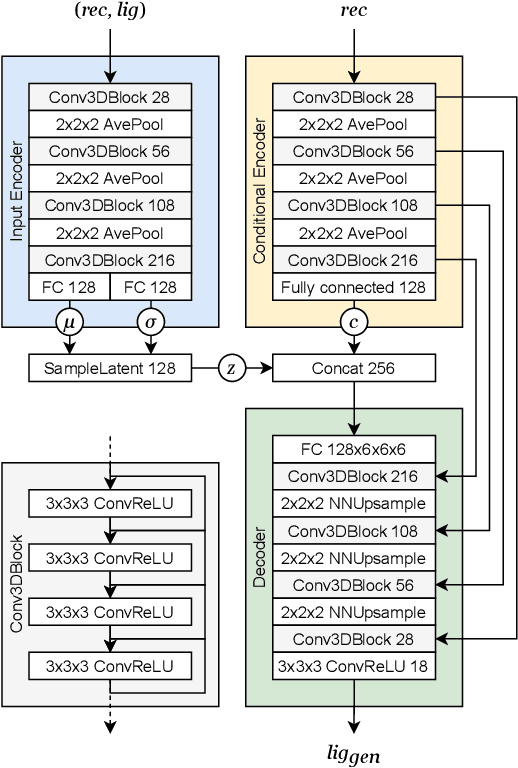
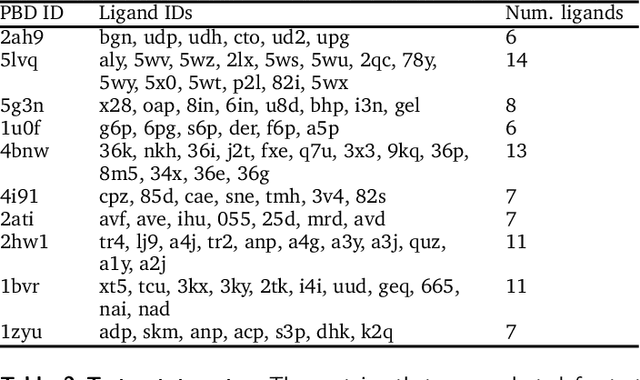
Abstract:The goal of structure-based drug discovery is to find small molecules that bind to a given target protein. Deep learning has been used to generate drug-like molecules with certain cheminformatic properties, but has not yet been applied to generating 3D molecules predicted to bind to proteins by sampling the conditional distribution of protein-ligand binding interactions. In this work, we describe for the first time a deep learning system for generating 3D molecular structures conditioned on a receptor binding site. We approach the problem using a conditional variational autoencoder trained on an atomic density grid representation of cross-docked protein-ligand structures. We apply atom fitting and bond inference procedures to construct valid molecular conformations from generated atomic densities. We evaluate the properties of the generated molecules and demonstrate that they change significantly when conditioned on mutated receptors. We also explore the latent space learned by our generative model using sampling and interpolation techniques. This work opens the door for end-to-end prediction of stable bioactive molecules from protein structures with deep learning.
Learning a Continuous Representation of 3D Molecular Structures with Deep Generative Models
Oct 20, 2020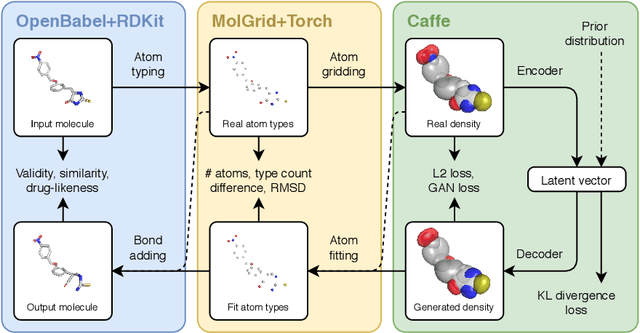
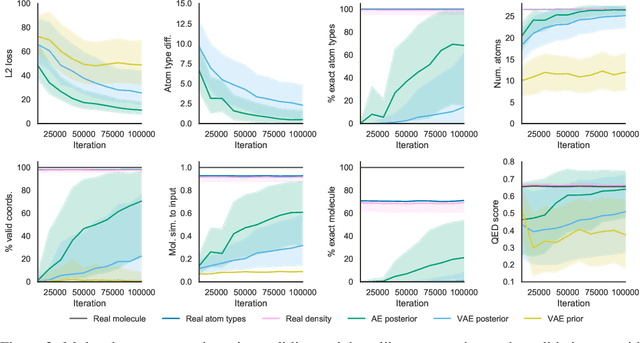
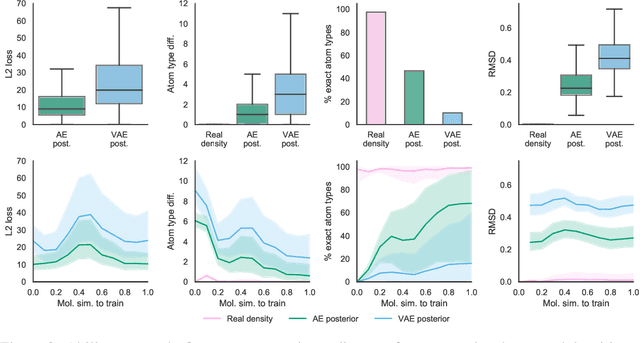
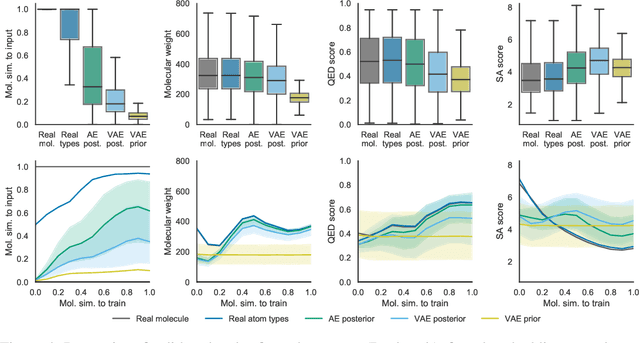
Abstract:Machine learning methods in drug discovery have primarily focused on virtual screening of molecular libraries using discriminative models. Generative models are an entirely different approach to drug discovery that learn to represent and optimize molecules in a continuous latent space. These methods have already been applied with increasing success to the generation of two dimensional molecules as SMILES strings and molecular graphs. In this work, we describe deep generative models for three dimensional molecular structures using atomic density grids and a novel fitting algorithm that converts continuous grids to discrete molecular structures. Our models jointly represent drug-like molecules and their conformations in a latent space that can be explored through interpolation. We are able to sample diverse sets of molecules based on a given input compound and increase the probability of creating a valid, drug-like molecule.
Generating 3D Molecular Structures Conditional on a Receptor Binding Site with Deep Generative Models
Oct 16, 2020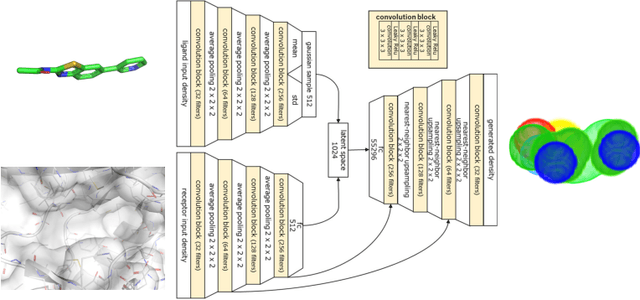
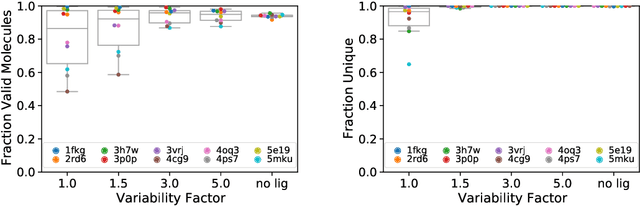
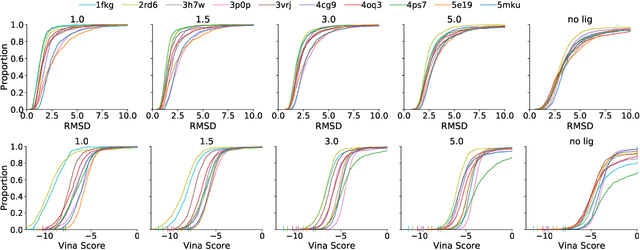
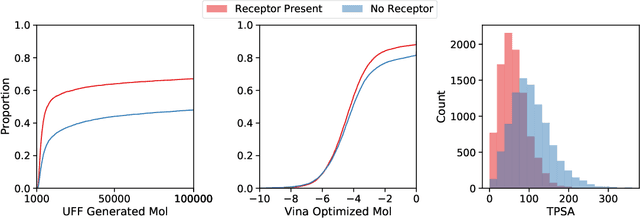
Abstract:Deep generative models have been applied with increasing success to the generation of two dimensional molecules as SMILES strings and molecular graphs. In this work we describe for the first time a deep generative model that can generate 3D molecular structures conditioned on a three-dimensional (3D) binding pocket. Using convolutional neural networks, we encode atomic density grids into separate receptor and ligand latent spaces. The ligand latent space is variational to support sampling of new molecules. A decoder network generates atomic densities of novel ligands conditioned on the receptor. Discrete atoms are then fit to these continuous densities to create molecular structures. We show that valid and unique molecules can be readily sampled from the variational latent space defined by a reference `seed' structure and generated structures have reasonable interactions with the binding site. As structures are sampled farther in latent space from the seed structure, the novelty of the generated structures increases, but the predicted binding affinity decreases. Overall, we demonstrate the feasibility of conditional 3D molecular structure generation and provide a starting point for methods that also explicitly optimize for desired molecular properties, such as high binding affinity.
Visualizing Convolutional Neural Network Protein-Ligand Scoring
Mar 06, 2018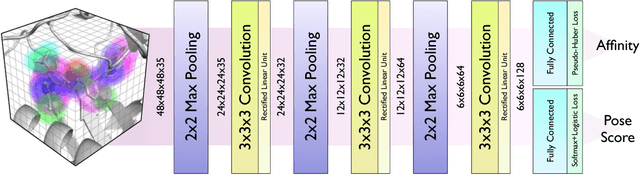

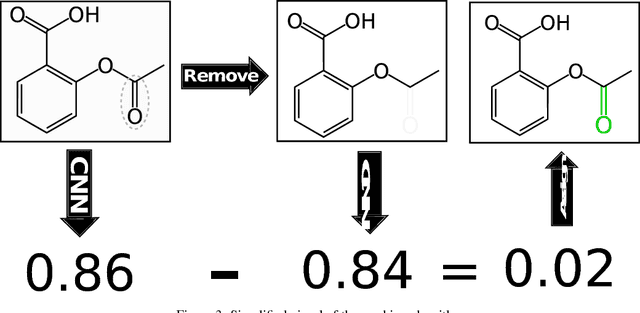
Abstract:Protein-ligand scoring is an important step in a structure-based drug design pipeline. Selecting a correct binding pose and predicting the binding affinity of a protein-ligand complex enables effective virtual screening. Machine learning techniques can make use of the increasing amounts of structural data that are becoming publicly available. Convolutional neural network (CNN) scoring functions in particular have shown promise in pose selection and affinity prediction for protein-ligand complexes. Neural networks are known for being difficult to interpret. Understanding the decisions of a particular network can help tune parameters and training data to maximize performance. Visualization of neural networks helps decompose complex scoring functions into pictures that are more easily parsed by humans. Here we present three methods for visualizing how individual protein-ligand complexes are interpreted by 3D convolutional neural networks. We also present a visualization of the convolutional filters and their weights. We describe how the intuition provided by these visualizations aids in network design.
Ligand Pose Optimization with Atomic Grid-Based Convolutional Neural Networks
Oct 20, 2017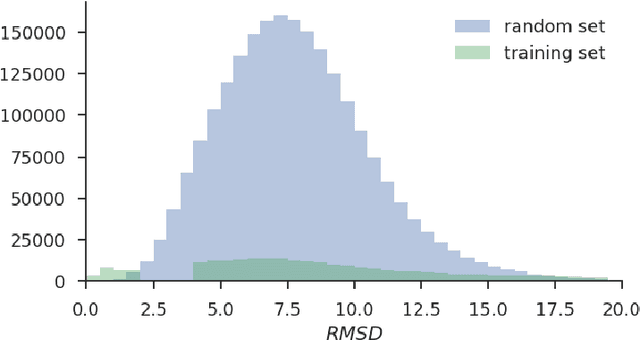

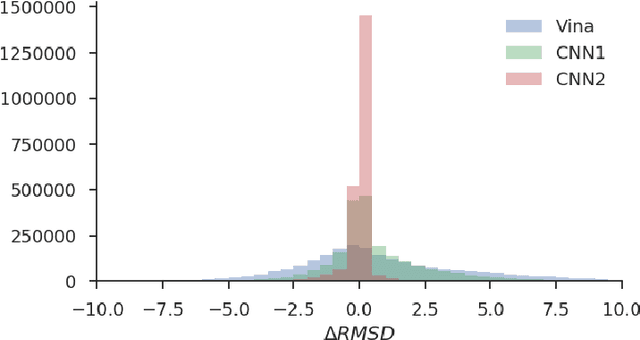
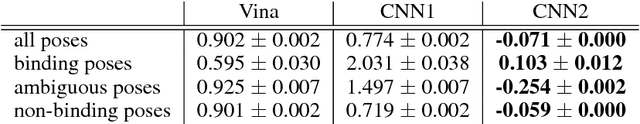
Abstract:Docking is an important tool in computational drug discovery that aims to predict the binding pose of a ligand to a target protein through a combination of pose scoring and optimization. A scoring function that is differentiable with respect to atom positions can be used for both scoring and gradient-based optimization of poses for docking. Using a differentiable grid-based atomic representation as input, we demonstrate that a scoring function learned by training a convolutional neural network (CNN) to identify binding poses can also be applied to pose optimization. We also show that an iteratively-trained CNN that includes poses optimized by the first CNN in its training set performs even better at optimizing randomly initialized poses than either the first CNN scoring function or AutoDock Vina.
Protein-Ligand Scoring with Convolutional Neural Networks
Dec 08, 2016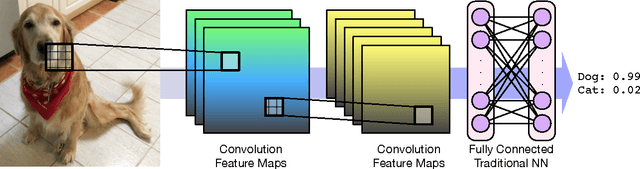
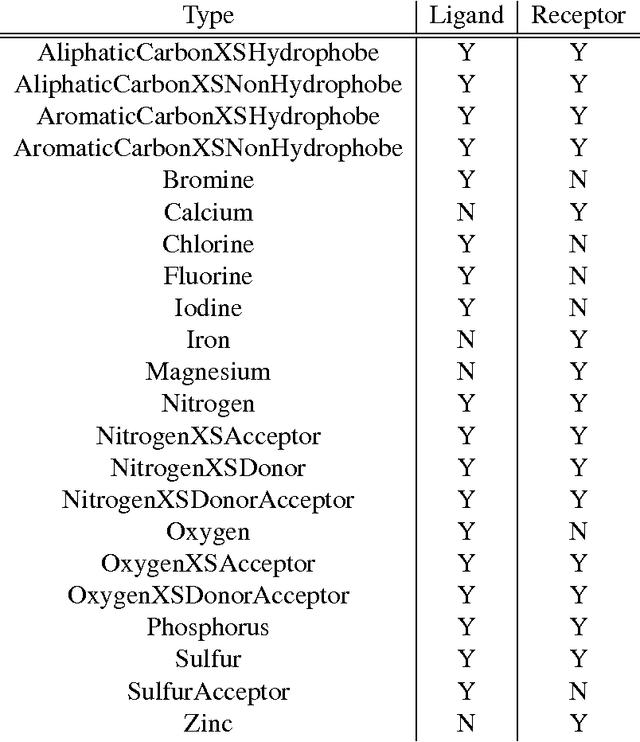
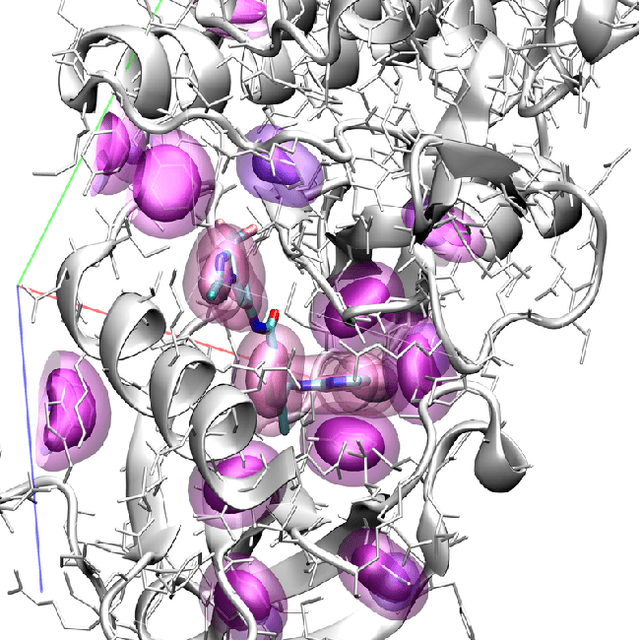
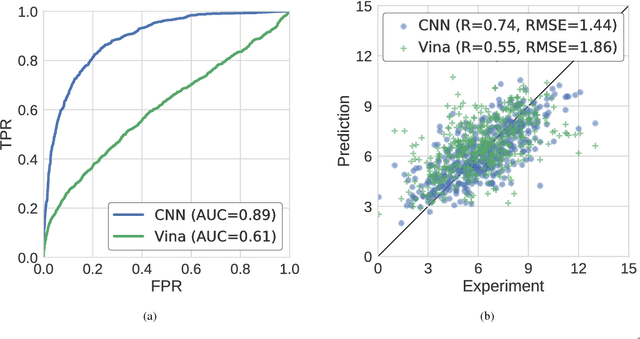
Abstract:Computational approaches to drug discovery can reduce the time and cost associated with experimental assays and enable the screening of novel chemotypes. Structure-based drug design methods rely on scoring functions to rank and predict binding affinities and poses. The ever-expanding amount of protein-ligand binding and structural data enables the use of deep machine learning techniques for protein-ligand scoring. We describe convolutional neural network (CNN) scoring functions that take as input a comprehensive 3D representation of a protein-ligand interaction. A CNN scoring function automatically learns the key features of protein-ligand interactions that correlate with binding. We train and optimize our CNN scoring functions to discriminate between correct and incorrect binding poses and known binders and non-binders. We find that our CNN scoring function outperforms the AutoDock Vina scoring function when ranking poses both for pose prediction and virtual screening.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge